
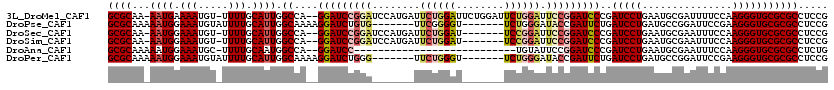
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,358,829 – 18,358,945 |
| Length | 116 |
| Max. P | 0.986732 |

| Location | 18,358,829 – 18,358,945 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.32 |
| Mean single sequence MFE | -42.08 |
| Consensus MFE | -25.38 |
| Energy contribution | -26.41 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
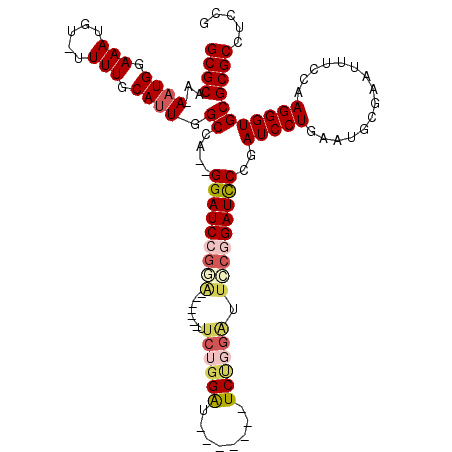
>3L_DroMel_CAF1 18358829 116 + 23771897 GCGCAA-AAUGAAAAUGU-UUUUGCAUUGGCCA--GGAUCCGGAUCCAUGAUUCUGGAUUCUGGAUUCUGGAUUCCGGAUCCCGAUCCUGAAUGCGAUUUUCCAAGGGUGCGCGCCUCCG (((((.-...(((((...-..(((((((...((--(((((.((((((..((.((..((........))..)).)).)))))).)))))))))))))))))))......)))))....... ( -45.50) >DroPse_CAF1 169086 106 + 1 GCGCAAAAAUGGAAAUGUAUUUUGCAUUGGCAAAAGGAUCUGUG-------UUCGGGGU-------UCUGGGAUACCGAUUCUGAUCCUGAUGCCGGAUUCCGAAGGGUGCGCGCCUCCG (((((....(((((((((.....)))))((((..((((((.(..-------.((((..(-------(....))..))))..).))))))..))))....)))).....)))))....... ( -38.90) >DroSec_CAF1 142893 109 + 1 GCGCAA-AAUGGAAAUGU-UUUUGCAUUGGCCA--GGAUCCGGAUCCAUGAUUCUGGAU-------UCCGGAUUCCGGAUCCCGAUCCUGAAUGCGAAUUUCCAAGGGUGCGCGCCUCCG (((((.-..(((((((..-..(((((((...((--(((((.((((((..((.(((((..-------.))))).)).)))))).)))))))))))))))))))))....)))))....... ( -50.60) >DroSim_CAF1 153408 109 + 1 GCGCAA-AAUGGAAAUGU-UUUUGCAUUGGCCA--GGAUCCGGAUCCAUGAUUCUGGAU-------UCCGGAUUCCGGAUCCCGAUCCUGAAUGCGAAUUUCCAAGGGUGCGCGCCUCCG (((((.-..(((((((..-..(((((((...((--(((((.((((((..((.(((((..-------.))))).)).)))))).)))))))))))))))))))))....)))))....... ( -50.60) >DroAna_CAF1 139390 90 + 1 GCGCAAAAAUGGAAAUGC-UUUUGCAAUGGCCA--GGAUCC---------------------------UGUAUUCCGGAUCCCGAUCCUGAAUGCGAAUUUCCAAGGGUGCGCGCCUCUG (((((....(((((((..-..(((((..((.(.--((((((---------------------------........)))))).)..))....))))))))))))....)))))....... ( -30.70) >DroPer_CAF1 170064 106 + 1 GCGCAAAAAUGGAAAUGUAUUUUGCAUUGGCAAAAGGAUCUGGG-------UUCUGGGU-------UCUGGGAUACCGAUUCUGAUCCUGAUGCCGGAUUCCGAAGGGUGCGCGCCUCCG (((((....(((((((((.....)))))((((..((((((.(((-------((...(((-------........)))))))).))))))..))))....)))).....)))))....... ( -36.20) >consensus GCGCAA_AAUGGAAAUGU_UUUUGCAUUGGCCA__GGAUCCGGA_______UUCUGGAU_______UCUGGAUUCCGGAUCCCGAUCCUGAAUGCGAAUUUCCAAGGGUGCGCGCCUCCG ((((...((((.(((.....))).)))).((....(((((((((........((((((........)))))).)))))))))..(((((...............)))))))))))..... (-25.38 = -26.41 + 1.03)
| Location | 18,358,829 – 18,358,945 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.32 |
| Mean single sequence MFE | -37.68 |
| Consensus MFE | -18.01 |
| Energy contribution | -18.98 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.974171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
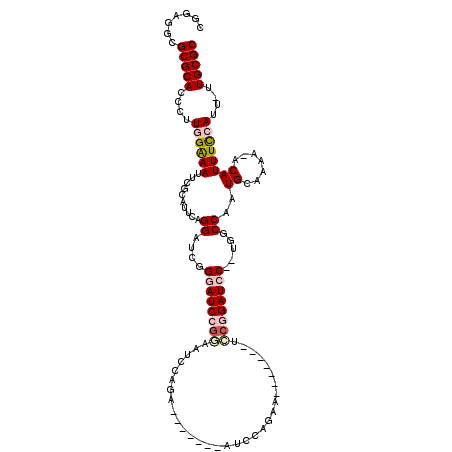
>3L_DroMel_CAF1 18358829 116 - 23771897 CGGAGGCGCGCACCCUUGGAAAAUCGCAUUCAGGAUCGGGAUCCGGAAUCCAGAAUCCAGAAUCCAGAAUCAUGGAUCCGGAUCC--UGGCCAAUGCAAAA-ACAUUUUCAUU-UUGCGC .......(((((....((((((...(((((..((.((((((((((((.((((((.((.........)).)).)))))))))))))--))))))))))....-...))))))..-.))))) ( -42.60) >DroPse_CAF1 169086 106 - 1 CGGAGGCGCGCACCCUUCGGAAUCCGGCAUCAGGAUCAGAAUCGGUAUCCCAGA-------ACCCCGAA-------CACAGAUCCUUUUGCCAAUGCAAAAUACAUUUCCAUUUUUGCGC ((((((.(....)))))))......((((..((((((....((((.........-------...)))).-------....))))))..))))...((((((...........)))))).. ( -30.50) >DroSec_CAF1 142893 109 - 1 CGGAGGCGCGCACCCUUGGAAAUUCGCAUUCAGGAUCGGGAUCCGGAAUCCGGA-------AUCCAGAAUCAUGGAUCCGGAUCC--UGGCCAAUGCAAAA-ACAUUUCCAUU-UUGCGC .......(((((....(((((((..(((((..((.((((((((((((.((((((-------........)).)))))))))))))--))))))))))....-..)))))))..-.))))) ( -45.50) >DroSim_CAF1 153408 109 - 1 CGGAGGCGCGCACCCUUGGAAAUUCGCAUUCAGGAUCGGGAUCCGGAAUCCGGA-------AUCCAGAAUCAUGGAUCCGGAUCC--UGGCCAAUGCAAAA-ACAUUUCCAUU-UUGCGC .......(((((....(((((((..(((((..((.((((((((((((.((((((-------........)).)))))))))))))--))))))))))....-..)))))))..-.))))) ( -45.50) >DroAna_CAF1 139390 90 - 1 CAGAGGCGCGCACCCUUGGAAAUUCGCAUUCAGGAUCGGGAUCCGGAAUACA---------------------------GGAUCC--UGGCCAUUGCAAAA-GCAUUUCCAUUUUUGCGC .......(((((....((((((..........((.(((((((((........---------------------------))))))--)))))..(((....-)))))))))....))))) ( -33.70) >DroPer_CAF1 170064 106 - 1 CGGAGGCGCGCACCCUUCGGAAUCCGGCAUCAGGAUCAGAAUCGGUAUCCCAGA-------ACCCAGAA-------CCCAGAUCCUUUUGCCAAUGCAAAAUACAUUUCCAUUUUUGCGC ((((((.(....)))))))......((((..((((((......(((........-------)))..(..-------..).))))))..))))...((((((...........)))))).. ( -28.30) >consensus CGGAGGCGCGCACCCUUGGAAAUUCGCAUUCAGGAUCGGGAUCCGGAAUCCAGA_______AUCCAGAA_______UCCGGAUCC__UGGCCAAUGCAAAA_ACAUUUCCAUU_UUGCGC .......(((((....((((((..........((....((((((((...............................)))))))).....))..((.......))))))))....))))) (-18.01 = -18.98 + 0.97)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:05 2006