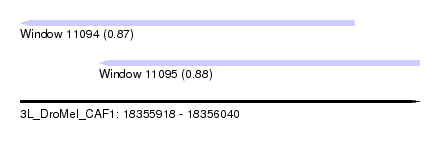
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,355,918 – 18,356,040 |
| Length | 122 |
| Max. P | 0.880608 |

| Location | 18,355,918 – 18,356,020 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 80.19 |
| Mean single sequence MFE | -24.35 |
| Consensus MFE | -17.65 |
| Energy contribution | -17.18 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
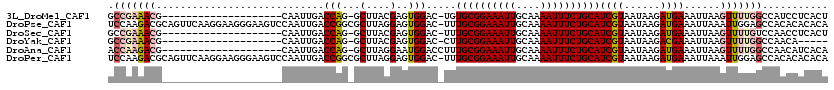
>3L_DroMel_CAF1 18355918 102 - 23771897 -GCUUACGAGUGGAC-UGUGCGGAAAUUGCAAAAUUUCUGCAUCGUAAUAAGAUGAAAUUAAGUUUUGGCCAUCCUCACUGAAGCCCACACUCC--UUUA----CAGUAC -......(((((.((-.(((((((((((....))))))))))).))................((((..(.........)..))))...))))).--....----...... ( -24.40) >DroPse_CAF1 165514 92 - 1 CGCUUAGGAGUGGAC-UUUGCGGAAAUUGCAAAAUUUCUGCAUCGUAAUAAGAUGAAAUUAAAUUGGAGCCACACACACACAGGCACACACGC----------------- .((((..(.((((.(-((((((((((((....))))))))).((((......)))).........)))))))).)......))))........----------------- ( -22.30) >DroSec_CAF1 140067 102 - 1 -GCUUACGAGUGGAC-UUUGCGGAAAUUGCAAAAUUUCUGCAUCGUAAUAAGAUGAAAUUAAGUUUUGUCCAACCUCACUGAAUCCCAUACUCC--UUCA----CAGCCC -(((...((((((((-..((((((((((....))))))))))((((......))))...........)))))..)))..((((...........--))))----.))).. ( -22.90) >DroEre_CAF1 135388 102 - 1 -GCUUACGAGUGGAC-CUUGCGGAAAUUGCAAAAUUUCUGCAUCGUAAUAAGAUGAAAUUAAGUUUUGGCCAACCUCACUGAUUCCCACACUCC--UUCG----CAGCAC -((..(.(((((.((-..((((((((((....))))))))))..))........(((....(((...((....))..)))..)))...))))).--)..)----)..... ( -23.10) >DroYak_CAF1 146236 96 - 1 -GCUUACGAGUGGAC-CUUGCGGAAAUUGCAAAAUUUCUGCAUCGUAAUAAGACGAAAUUAAGUUUUGGCCAACA------CUUCCCACACUCC--UCUA----CAGCAC -(((...(((((.((-..((((((((((....))))))))))..))..((((((........)))))).......------.......))))).--....----.))).. ( -24.50) >DroAna_CAF1 136508 109 - 1 -GCUUAGGAAUGGACCUUUGCGGAAAUUGCAAAAUUUCUGCAUCGUAAUAAGAUGAAAUUAAGUUUUGGCCAACAUCACACAUUCCCGUACACACGUACGAACUCAAAGC -((((.((((((......((((((((((....))))))))))((((......))))......((..((.....))..)).))))))(((((....)))))......)))) ( -28.90) >consensus _GCUUACGAGUGGAC_UUUGCGGAAAUUGCAAAAUUUCUGCAUCGUAAUAAGAUGAAAUUAAGUUUUGGCCAACCUCACUGAUUCCCACACUCC__UUCA____CAGCAC .........((((.....((((((((((....))))))))))((((......)))).............................))))..................... (-17.65 = -17.18 + -0.47)



| Location | 18,355,942 – 18,356,040 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.57 |
| Mean single sequence MFE | -29.48 |
| Consensus MFE | -15.99 |
| Energy contribution | -16.80 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18355942 98 - 23771897 GCCGAAACG--------------------CAAUUGACCAG-GCUUACGAGUGGAC-UGUGCGGAAAUUGCAAAAUUUCUGCAUCGUAAUAAGAUGAAAUUAAGUUUUGGCCAUCCUCACU ((((((((.--------------------.((((..(((.-.(....)..)))((-.(((((((((((....))))))))))).))..........))))..)))))))).......... ( -28.90) >DroPse_CAF1 165527 119 - 1 UCCAAGACGCAGUUCAAGGAAGGGAAGUCCAAUUGACCGGCGCUUAGGAGUGGAC-UUUGCGGAAAUUGCAAAAUUUCUGCAUCGUAAUAAGAUGAAAUUAAAUUGGAGCCACACACACA ((((((.(((.(.((((.....((....))..)))).).)))))).)))((((.(-((((((((((((....))))))))).((((......)))).........))))))))....... ( -34.60) >DroSec_CAF1 140091 98 - 1 GCCGAAACG--------------------CAAUUGACCAG-GCUUACGAGUGGAC-UUUGCGGAAAUUGCAAAAUUUCUGCAUCGUAAUAAGAUGAAAUUAAGUUUUGUCCAACCUCACU (((......--------------------..........)-))....((((((((-..((((((((((....))))))))))((((......))))...........)))))..)))... ( -25.19) >DroYak_CAF1 146259 93 - 1 GCCGAAACG--------------------CAAUUGACCAG-GCUUACGAGUGGAC-CUUGCGGAAAUUGCAAAAUUUCUGCAUCGUAAUAAGACGAAAUUAAGUUUUGGCCAACA----- ((((((((.--------------------.((((..(((.-.(....)..)))..-..((((((((((....))))))))))((((......))))))))..)))))))).....----- ( -28.50) >DroAna_CAF1 136538 99 - 1 ACCAAGACG--------------------CAAUUGACCAG-GCUUAGGAAUGGACCUUUGCGGAAAUUGCAAAAUUUCUGCAUCGUAAUAAGAUGAAAUUAAGUUUUGGCCAACAUCACA .((((((((--------------------(((((..((..-((..(((......)))..)))).))))))..........((((.......)))).......)))))))........... ( -25.10) >DroPer_CAF1 166544 119 - 1 UCCAAGACGCAGUUCAAGGAAGGGAAGUCCAAUUGACCGGCGCUUAGGAGUGGAC-UUUGCGGAAAUUGCAAAAUUUCUGCAUCGUAAUAAGAUGAAAUUAAAUUGGAGCCACACACACA ((((((.(((.(.((((.....((....))..)))).).)))))).)))((((.(-((((((((((((....))))))))).((((......)))).........))))))))....... ( -34.60) >consensus GCCAAAACG____________________CAAUUGACCAG_GCUUACGAGUGGAC_UUUGCGGAAAUUGCAAAAUUUCUGCAUCGUAAUAAGAUGAAAUUAAGUUUUGGCCAACCUCACA .(((((((............................(((...(....)..))).....((((((((((....))))))))))((((......))))......)))))))........... (-15.99 = -16.80 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:03 2006