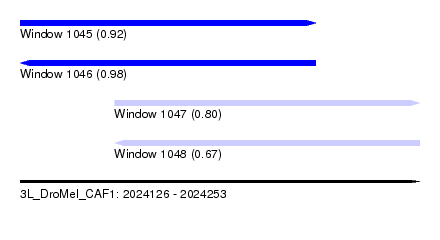
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,024,126 – 2,024,253 |
| Length | 127 |
| Max. P | 0.981602 |

| Location | 2,024,126 – 2,024,220 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 94.39 |
| Mean single sequence MFE | -23.83 |
| Consensus MFE | -23.66 |
| Energy contribution | -23.00 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
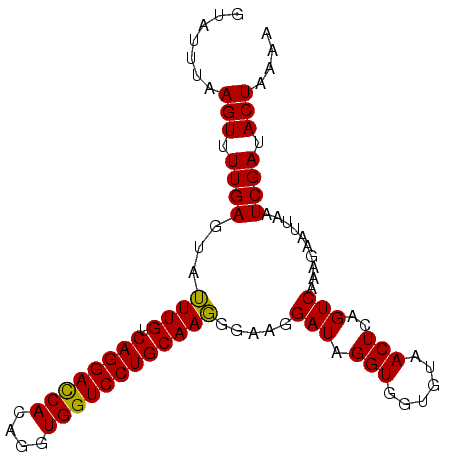
>3L_DroMel_CAF1 2024126 94 + 23771897 GAAUGUAAGUUUUGAGUAAUUGUCAGGAUCACAGGUGGUCCUGCAAUGG-AGGAUAGGUGGUGUAACUCAGUCGAAGAAUUAAUCGACACUAAAA .......(((..(((((.((((.((((((((....))))))))))))..-...(((.....))).)))))(((((........)))))))).... ( -23.90) >DroSec_CAF1 74585 95 + 1 GUAUUUAAGUUUUGAGUAUUUGUCAGGACCACAGGUGGUCCUGCAAGGGAAGGAUAGGUGGUGUAACUCAGUCAAAGAAUUAAUCGAUACUAAAA .......(((.((((...((((.((((((((....)))))))))))).....(((.(((......)))..)))..........)))).))).... ( -23.80) >DroSim_CAF1 89736 95 + 1 GUAUUUAAGUUUUGAGUAUUUGUCAGGACCACAGGUGGUCCUGCAAGGGAAGGAUAGGUGGUGUAACUCAGUCAAAGAAUUAAUCGAUACUAAAA .......(((.((((...((((.((((((((....)))))))))))).....(((.(((......)))..)))..........)))).))).... ( -23.80) >consensus GUAUUUAAGUUUUGAGUAUUUGUCAGGACCACAGGUGGUCCUGCAAGGGAAGGAUAGGUGGUGUAACUCAGUCAAAGAAUUAAUCGAUACUAAAA .......(((.((((...((((.((((((((....)))))))))))).....(((.(((......)))..)))..........)))).))).... (-23.66 = -23.00 + -0.66)

| Location | 2,024,126 – 2,024,220 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 94.39 |
| Mean single sequence MFE | -20.79 |
| Consensus MFE | -19.02 |
| Energy contribution | -19.13 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.66 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2024126 94 - 23771897 UUUUAGUGUCGAUUAAUUCUUCGACUGAGUUACACCACCUAUCCU-CCAUUGCAGGACCACCUGUGAUCCUGACAAUUACUCAAAACUUACAUUC .......(((((........)))))(((((...............-..(((((((((.((....)).))))).)))).)))))............ ( -18.97) >DroSec_CAF1 74585 95 - 1 UUUUAGUAUCGAUUAAUUCUUUGACUGAGUUACACCACCUAUCCUUCCCUUGCAGGACCACCUGUGGUCCUGACAAAUACUCAAAACUUAAAUAC (((.(((((....((((((.......)))))).................(((((((((((....)))))))).)))))))).))).......... ( -21.70) >DroSim_CAF1 89736 95 - 1 UUUUAGUAUCGAUUAAUUCUUUGACUGAGUUACACCACCUAUCCUUCCCUUGCAGGACCACCUGUGGUCCUGACAAAUACUCAAAACUUAAAUAC (((.(((((....((((((.......)))))).................(((((((((((....)))))))).)))))))).))).......... ( -21.70) >consensus UUUUAGUAUCGAUUAAUUCUUUGACUGAGUUACACCACCUAUCCUUCCCUUGCAGGACCACCUGUGGUCCUGACAAAUACUCAAAACUUAAAUAC (((.((((.....((((((.......)))))).................(((((((((((....)))))))).))).)))).))).......... (-19.02 = -19.13 + 0.11)

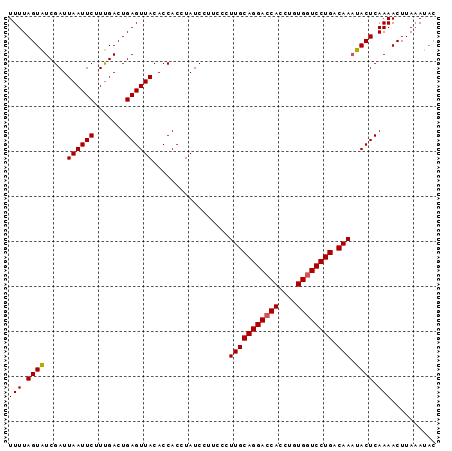
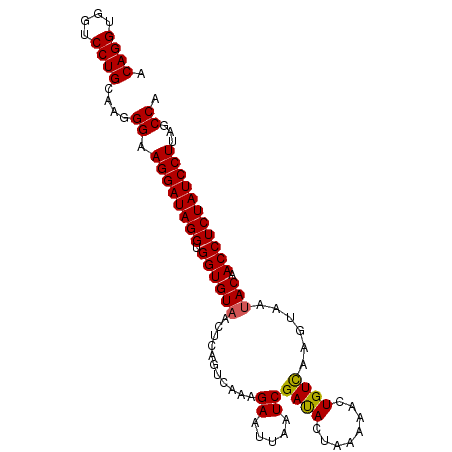
| Location | 2,024,156 – 2,024,253 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 94.56 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -22.17 |
| Energy contribution | -22.07 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
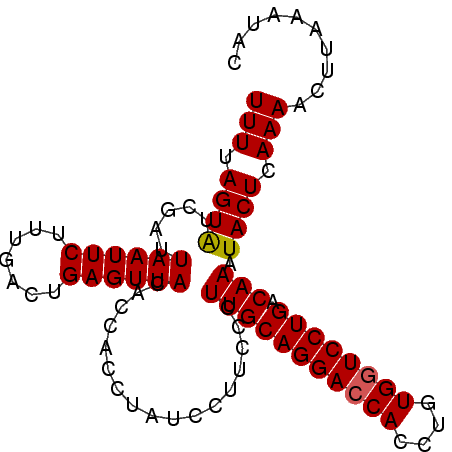
>3L_DroMel_CAF1 2024156 97 + 23771897 ACAGGUGGUCCUGCAAUGG-AGGAUAGGUGGUGUAACUCAGUCGAAGAAUUAAUCGACACUAAAAACUGUUAAAUAAUACAACCUCUAUCCUUAGCCA .((((....)))).....(-((((((((.((((((.....(((((........)))))((........)).......))).))))))))))))..... ( -26.10) >DroSec_CAF1 74615 98 + 1 ACAGGUGGUCCUGCAAGGGAAGGAUAGGUGGUGUAACUCAGUCAAAGAAUUAAUCGAUACUAAAAACUGUCAAGUAAUACAACCUCUAUCCUUAGCCA .((((....))))...((.(((((((((.(((((((((((((...((.(((....))).))....))))...)))..))).))))))))))))..)). ( -25.40) >DroSim_CAF1 89766 98 + 1 ACAGGUGGUCCUGCAAGGGAAGGAUAGGUGGUGUAACUCAGUCAAAGAAUUAAUCGAUACUAAAAAGUGUCAAGUAAGACAACCUCUAUCCUUAGCCA .((((....))))...((.(((((((((.((((.....).(((...((.....))((((((....))))))......))).))))))))))))..)). ( -28.70) >consensus ACAGGUGGUCCUGCAAGGGAAGGAUAGGUGGUGUAACUCAGUCAAAGAAUUAAUCGAUACUAAAAACUGUCAAGUAAUACAACCUCUAUCCUUAGCCA .((((....))))....((.((((((((.((((((...........((.....))((((........))))......))).)))))))))))...)). (-22.17 = -22.07 + -0.11)

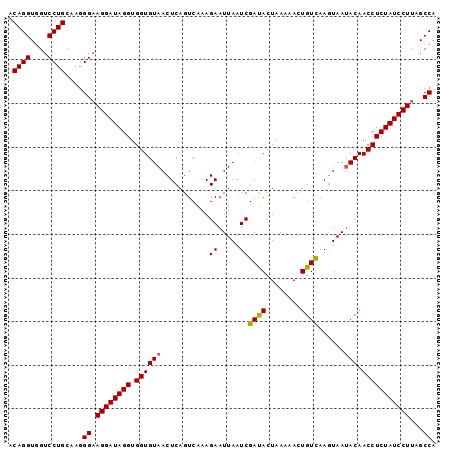
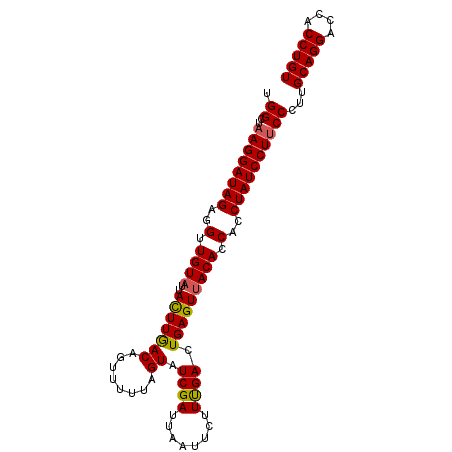
| Location | 2,024,156 – 2,024,253 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 94.56 |
| Mean single sequence MFE | -25.13 |
| Consensus MFE | -21.46 |
| Energy contribution | -21.47 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2024156 97 - 23771897 UGGCUAAGGAUAGAGGUUGUAUUAUUUAACAGUUUUUAGUGUCGAUUAAUUCUUCGACUGAGUUACACCACCUAUCCU-CCAUUGCAGGACCACCUGU (((...(((((((..(.((((..(((((((........))(((((........)))))))))))))).)..)))))))-)))..(((((....))))) ( -25.80) >DroSec_CAF1 74615 98 - 1 UGGCUAAGGAUAGAGGUUGUAUUACUUGACAGUUUUUAGUAUCGAUUAAUUCUUUGACUGAGUUACACCACCUAUCCUUCCCUUGCAGGACCACCUGU .((..((((((((..(.((((..(((...(((((...((............))..)))))))))))).)..)))))))).))..(((((....))))) ( -24.50) >DroSim_CAF1 89766 98 - 1 UGGCUAAGGAUAGAGGUUGUCUUACUUGACACUUUUUAGUAUCGAUUAAUUCUUUGACUGAGUUACACCACCUAUCCUUCCCUUGCAGGACCACCUGU .((..((((((((.(((((((......))))...((((((...((.....))....))))))....)))..)))))))).))..(((((....))))) ( -25.10) >consensus UGGCUAAGGAUAGAGGUUGUAUUACUUGACAGUUUUUAGUAUCGAUUAAUUCUUUGACUGAGUUACACCACCUAUCCUUCCCUUGCAGGACCACCUGU .((..((((((((..(.((((..(((((((........)).((((........)))).))))))))).)..))))))))))...(((((....))))) (-21.46 = -21.47 + 0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:29 2006