| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,346,046 – 18,346,149 |
| Length | 103 |
| Max. P | 0.657927 |

| Location | 18,346,046 – 18,346,149 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.59 |
| Mean single sequence MFE | -26.27 |
| Consensus MFE | -14.09 |
| Energy contribution | -14.57 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
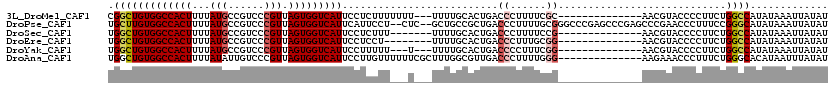
>3L_DroMel_CAF1 18346046 103 + 23771897 CGGCUGUGGCCACUUUUAUGCCGUCCCGUUAGUGGUCAUUCCUCUUUUUUU---UUUUGCACUGACCCUUUUCGC--------------AACGUACCCCUUCUGGCCAUAUAAAUUAUAU ....((((((((.........(((..((..((.(((((.............---........)))))))...)).--------------.))).........)))))))).......... ( -21.57) >DroPse_CAF1 156235 116 + 1 UGCUUGUGGCCACUUUUAUGCCGUCCCGUUAGUGGUCAUUCAUUCCU--CUC--GCUGCCGCUGACCCUUUUGCGGGCCCGAGCCCGAGCCCGAACCCUUUCCGGGCAUAUAAAUUAUAU .(((((.((((.......(((......(((((((((...........--...--...)))))))))......))))))))))))....(((((.........)))))............. ( -34.14) >DroSec_CAF1 130415 99 + 1 UGGCUGUGGCCACUUUUAUGCCGUCCCGUUAGUGGUCAUUCCUCUUU-------UUUUGCACUGACCCUUUUCCG--------------AACGUACCCCUUCUGGCCAUAUAAAUUAUAU ....((((((((.........(((.(.(..((.(((((.........-------........)))))))...).)--------------.))).........)))))))).......... ( -19.30) >DroEre_CAF1 125933 98 + 1 UGGCUGUGGCCACUUUUAUGCCGUCCCGUUAGUGGUCAUUCCUCCU--------UUUUGCACUGACCCUUUGCGG--------------AACGUACCCCUUCUGGCCAUAUAAAUUAUAU ....((((((((.........(((.((((.((.(((((...(....--------....)...)))))))..))))--------------.))).........)))))))).......... ( -27.27) >DroYak_CAF1 136805 100 + 1 UGGCUGUGGCCACUUUUAUGCCGUCCCGUUAGUGGUCAUUCCUUUUU---U---UUUUGCACUGACCCCUUUCGG--------------AACGUACCCCUUCUGGCCAUAUAAAUUAUAU ....((((((((.........(((.(((..((.(((((.........---.---........))))).))..)))--------------.))).........)))))))).......... ( -26.64) >DroAna_CAF1 126641 106 + 1 UGGCUGUGGCCACUUUUAUAUUGUCCCGUUAGUGGUCAUUCCUUGUUUUUUCGCUUUGGCGUUGACCCUUUUGGG--------------AAGAAACCCUUUCUGGGCACAUAAUUUAUAU ....((((.((............(((((..((.(((((..((..((......))...))...)))))))..))))--------------)((((.....)))))).)))).......... ( -28.70) >consensus UGGCUGUGGCCACUUUUAUGCCGUCCCGUUAGUGGUCAUUCCUCUUU___U___UUUUGCACUGACCCUUUUCGG______________AACGUACCCCUUCUGGCCAUAUAAAUUAUAU .(((((((((((((...(((......))).)))))))))..........................((......))............................))))............. (-14.09 = -14.57 + 0.47)
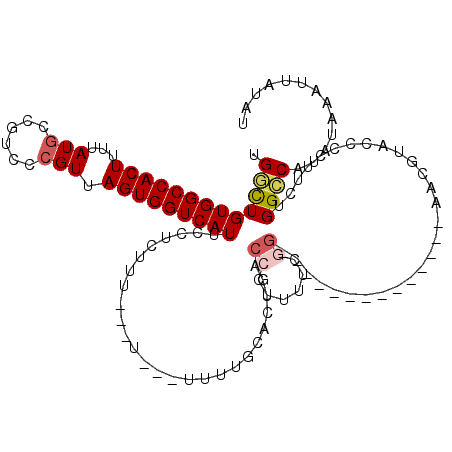
| Location | 18,346,046 – 18,346,149 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.59 |
| Mean single sequence MFE | -28.23 |
| Consensus MFE | -13.52 |
| Energy contribution | -13.66 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.48 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18346046 103 - 23771897 AUAUAAUUUAUAUGGCCAGAAGGGGUACGUU--------------GCGAAAAGGGUCAGUGCAAAA---AAAAAAAGAGGAAUGACCACUAACGGGACGGCAUAAAAGUGGCCACAGCCG ............((((((......((.((((--------------.((...(((((((.(.(....---.........).).))))).))..)).)))))).......))))))...... ( -23.74) >DroPse_CAF1 156235 116 - 1 AUAUAAUUUAUAUGCCCGGAAAGGGUUCGGGCUCGGGCUCGGGCCCGCAAAAGGGUCAGCGGCAGC--GAG--AGGAAUGAAUGACCACUAACGGGACGGCAUAAAAGUGGCCACAAGCA ..............((((....((((((((((....)))))))))).....(((((((....((.(--...--.)...))..))))).))..))))..(((.........)))....... ( -38.80) >DroSec_CAF1 130415 99 - 1 AUAUAAUUUAUAUGGCCAGAAGGGGUACGUU--------------CGGAAAAGGGUCAGUGCAAAA-------AAAGAGGAAUGACCACUAACGGGACGGCAUAAAAGUGGCCACAGCCA ............((((((......((.((((--------------(.(...(((((((.(.(....-------.....).).))))).))..).))))))).......))))))...... ( -23.62) >DroEre_CAF1 125933 98 - 1 AUAUAAUUUAUAUGGCCAGAAGGGGUACGUU--------------CCGCAAAGGGUCAGUGCAAAA--------AGGAGGAAUGACCACUAACGGGACGGCAUAAAAGUGGCCACAGCCA ............((((((......((.((((--------------(((...(((((((.(.(....--------....).).))))).))..))))))))).......))))))...... ( -28.72) >DroYak_CAF1 136805 100 - 1 AUAUAAUUUAUAUGGCCAGAAGGGGUACGUU--------------CCGAAAGGGGUCAGUGCAAAA---A---AAAAAGGAAUGACCACUAACGGGACGGCAUAAAAGUGGCCACAGCCA ............((((((......((.((((--------------(((..((.(((((.(.(....---.---.....).).))))).))..))))))))).......))))))...... ( -28.82) >DroAna_CAF1 126641 106 - 1 AUAUAAAUUAUGUGCCCAGAAAGGGUUUCUU--------------CCCAAAAGGGUCAACGCCAAAGCGAAAAAACAAGGAAUGACCACUAACGGGACAAUAUAAAAGUGGCCACAGCCA .......((((((.(((.....(((......--------------)))...(((((((...((...............))..))))).))...)))...))))))...((((....)))) ( -25.66) >consensus AUAUAAUUUAUAUGGCCAGAAGGGGUACGUU______________CCGAAAAGGGUCAGUGCAAAA___A___AAAAAGGAAUGACCACUAACGGGACGGCAUAAAAGUGGCCACAGCCA ............((((((......((.((((......................(((((........................))))).(....).)))))).......))))))...... (-13.52 = -13.66 + 0.14)

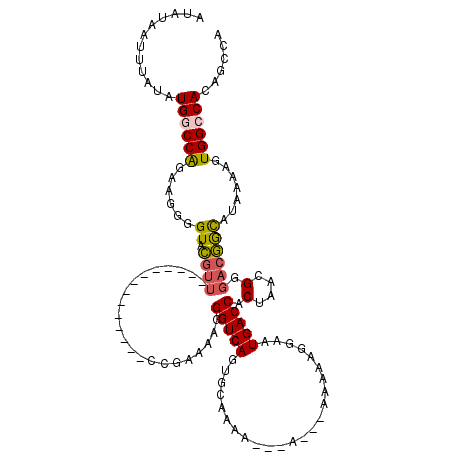
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:31:00 2006