
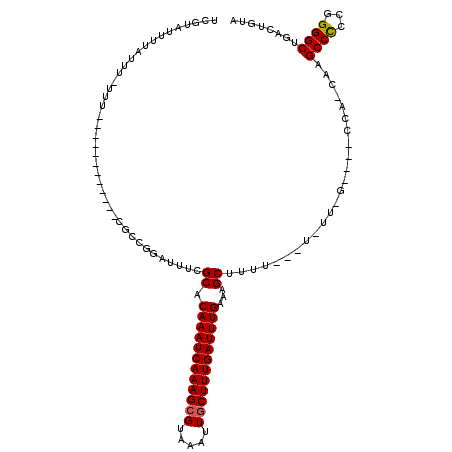
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,341,737 – 18,341,870 |
| Length | 133 |
| Max. P | 0.971053 |

| Location | 18,341,737 – 18,341,838 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.35 |
| Mean single sequence MFE | -25.05 |
| Consensus MFE | -20.20 |
| Energy contribution | -20.15 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
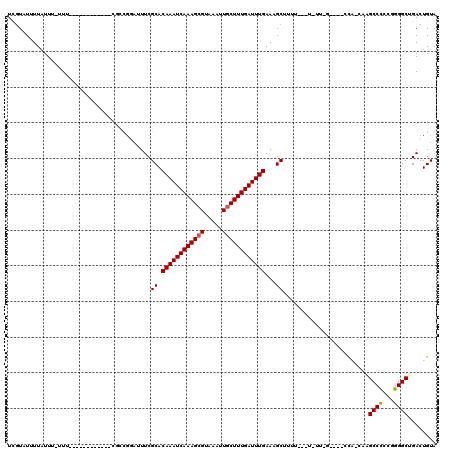
>3L_DroMel_CAF1 18341737 101 + 23771897 UUGUGUUUUAUUU-UUU------------CGCCGGAUUUCGCACAAAUCAAAGCGAAAAUUGCUUUGAUUUGAAAGCUUUUUUUUUUU-G----CCA-CAAGCCCCCGGGGCAGACUGUA (((((........-...------------.((........)).(((((((((((((...)))))))))))))................-.----.))-)))((((...))))........ ( -25.70) >DroSec_CAF1 126129 98 + 1 UUGUAUUUUAUUU-UUU------------CGCCGGAUUUCGCACAAAUCAAAGCGAAAAUUGCUUUGAUUUGAAAGCUUUU---UUUU-G----CCA-CAAGCCCGCGGGGCAGACUGUA .............-(..------------(((.((........(((((((((((((...)))))))))))))...((....---....-)----)..-....)).)))..)......... ( -24.80) >DroEre_CAF1 121759 97 + 1 UCGUAUUUUAUUU--UU------------CGCCGGAUUUCGCACAAAUCAAAGCGUAAAUUGCUUUGAUUUGAAAGCUUUU---UGUU-G----CCA-CAAGCCCCCGGGGCUGACUGUA .............--..------------...(((.....((.((((((((((((.....))))))))))))..(((....---.)))-)----)..-..(((((...)))))..))).. ( -25.20) >DroYak_CAF1 132481 97 + 1 UCGUAUUUUAUUU-UUU------------CGUCGGAUUUCGCACAAAUCAAAGCGUAAAUUUCUUUGAUUUGAAAGCAUAU---U-UU-G----CCA-CAAGCCCCCGGGGCUGACUGUA .............-...------------.(((((.(..((..((((((((((.(.....).))))))))))...(((...---.-.)-)----)..-........))..)))))).... ( -21.70) >DroAna_CAF1 122896 87 + 1 UGGUUUUUUAUUU-UU---------------------UUCGCACAAAUCAAAGCGUAAAUUGCUUUGAUUUGAAAGCUUUU---U-AU-G----CG--AAGGCCUCCAGGGCUGACUGUA .((((........-..---------------------((((((((((((((((((.....)))))))))))).........---.-.)-)----))--))((((.....))))))))... ( -26.60) >DroPer_CAF1 153300 117 + 1 UCUUUUUUUUUUUUUUUGGUUUUUUAUGCCUCUGCAAUUCGCACAAAUCAAAGCGUAAAUUGCUUUGAUUUGAAAGCUUUU---UAUUUGAAUGUUUUAAGGCCUUCCGGGCUGACUGUA ...............(..((((.....((((.(((.....)))((((((((((((.....))))))))))))(((((.(((---.....))).))))).)))).....))))..)..... ( -26.30) >consensus UCGUAUUUUAUUU_UUU____________CGCCGGAUUUCGCACAAAUCAAAGCGUAAAUUGCUUUGAUUUGAAAGCUUUU___U_UU_G____CCA_CAAGCCCCCGGGGCUGACUGUA ........................................((.((((((((((((.....))))))))))))...))........................((((...))))........ (-20.20 = -20.15 + -0.05)

| Location | 18,341,737 – 18,341,838 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.35 |
| Mean single sequence MFE | -24.93 |
| Consensus MFE | -20.08 |
| Energy contribution | -20.25 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18341737 101 - 23771897 UACAGUCUGCCCCGGGGGCUUG-UGG----C-AAAAAAAAAAAGCUUUCAAAUCAAAGCAAUUUUCGCUUUGAUUUGUGCGAAAUCCGGCG------------AAA-AAAUAAAACACAA ....(((((((.(((....)))-.))----)-)..........((...(((((((((((.......))))))))))).)).......))).------------...-............. ( -25.30) >DroSec_CAF1 126129 98 - 1 UACAGUCUGCCCCGCGGGCUUG-UGG----C-AAAA---AAAAGCUUUCAAAUCAAAGCAAUUUUCGCUUUGAUUUGUGCGAAAUCCGGCG------------AAA-AAAUAAAAUACAA ((((((((((...)))))).))-))(----(-....---....((...(((((((((((.......))))))))))).))........)).------------...-............. ( -27.89) >DroEre_CAF1 121759 97 - 1 UACAGUCAGCCCCGGGGGCUUG-UGG----C-AACA---AAAAGCUUUCAAAUCAAAGCAAUUUACGCUUUGAUUUGUGCGAAAUCCGGCG------------AA--AAAUAAAAUACGA ........(((..(..(((((.-((.----.-..))---..)))))..)((((((((((.......))))))))))...........))).------------..--............. ( -28.10) >DroYak_CAF1 132481 97 - 1 UACAGUCAGCCCCGGGGGCUUG-UGG----C-AA-A---AUAUGCUUUCAAAUCAAAGAAAUUUACGCUUUGAUUUGUGCGAAAUCCGACG------------AAA-AAAUAAAAUACGA ....((((((((...)))))..-...----.-..-.---...(((...((((((((((.........)))))))))).)))......))).------------...-............. ( -22.60) >DroAna_CAF1 122896 87 - 1 UACAGUCAGCCCUGGAGGCCUU--CG----C-AU-A---AAAAGCUUUCAAAUCAAAGCAAUUUACGCUUUGAUUUGUGCGAA---------------------AA-AAAUAAAAAACCA ........(((.....))).((--((----(-(.-.---.........(((((((((((.......)))))))))))))))))---------------------..-............. ( -21.60) >DroPer_CAF1 153300 117 - 1 UACAGUCAGCCCGGAAGGCCUUAAAACAUUCAAAUA---AAAAGCUUUCAAAUCAAAGCAAUUUACGCUUUGAUUUGUGCGAAUUGCAGAGGCAUAAAAAACCAAAAAAAAAAAAAAAGA ....(((((.((....)).))...............---....((...(((((((((((.......))))))))))).))..........)))........................... ( -24.10) >consensus UACAGUCAGCCCCGGGGGCUUG_UGG____C_AA_A___AAAAGCUUUCAAAUCAAAGCAAUUUACGCUUUGAUUUGUGCGAAAUCCGGCG____________AAA_AAAUAAAAUACGA .....((.(((.....)))........................((...(((((((((((.......))))))))))).))))...................................... (-20.08 = -20.25 + 0.17)

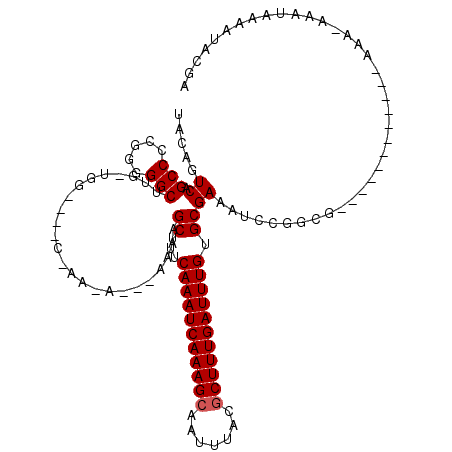

| Location | 18,341,764 – 18,341,870 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 81.35 |
| Mean single sequence MFE | -29.53 |
| Consensus MFE | -21.35 |
| Energy contribution | -21.38 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18341764 106 + 23771897 GCACAAAUCAAAGCGAAAAUUGCUUUGAUUUGAAAGCUUUUUUUUUUUGCCACAAGCCCCCGGGGCAGACUGUACCUCUCAGCCAAAUGAACGAA--AGGAAUUGGAG ...(((((((((((((...)))))))))))))....((..(((((((((......((((...)))).(.(((.......))).).......))))--)))))..)).. ( -29.10) >DroSec_CAF1 126156 93 + 1 GCACAAAUCAAAGCGAAAAUUGCUUUGAUUUGAAAGCUUUU---UUUUGCCACAAGCCCGCGGGGCAGACUGUACCUCUCAGCCA------------AAGAAGCGGAG ...(((((((((((((...)))))))))))))...((((((---((.........((((...)))).(.(((.......))).))------------))))))).... ( -29.70) >DroSim_CAF1 136365 104 + 1 GCACAAAUCAAAGCGAAAAUUGCUUUGAUUUGAAAGCUUUU--UUUUUGCCACAAGCCCGCGGGGCAGACUGUACCUCUCAGCGAAAUGAACGAA--AAGAAGCGGAG ...(((((((((((((...)))))))))))))...((((((--((((........((((...)))).(.(((.......)))).........)))--))))))).... ( -31.90) >DroEre_CAF1 121785 103 + 1 GCACAAAUCAAAGCGUAAAUUGCUUUGAUUUGAAAGCUUUU---UGUUGCCACAAGCCCCCGGGGCUGACUGUACCUCUCAGCCAAAUGAACGCA--GAGAAGUGGAG ((.((((((((((((.....))))))))))))...))....---.....((((.(((((...))))).........((((.((.........)).--)))).)))).. ( -32.80) >DroYak_CAF1 132508 104 + 1 GCACAAAUCAAAGCGUAAAUUUCUUUGAUUUGAAAGCAUAU---U-UUGCCACAAGCCCCCGGGGCUGACUGUACCUUUCAGCCAAAUGAACGAGAGUAGAAGUGGAG ((.((((((((((.(.....).))))))))))...))....---.-...((((.(((((...)))))...(.(((((((((......)))..))).))).).)))).. ( -26.90) >DroAna_CAF1 122914 83 + 1 GCACAAAUCAAAGCGUAAAUUGCUUUGAUUUGAAAGCUUUU---U-AUGCG-AAGGCCUCCAGGGCUGACUGUACCCGCCAGCCAAAU-------------------- ...((((((((((((.....))))))))))))...((((((---.-....)-)))))......(((((.(.......).)))))....-------------------- ( -26.80) >consensus GCACAAAUCAAAGCGAAAAUUGCUUUGAUUUGAAAGCUUUU___UUUUGCCACAAGCCCCCGGGGCAGACUGUACCUCUCAGCCAAAUGAACGAA__AAGAAGUGGAG ((.((((((((((((.....)))))))))))).......................((((...))))...............))......................... (-21.35 = -21.38 + 0.03)

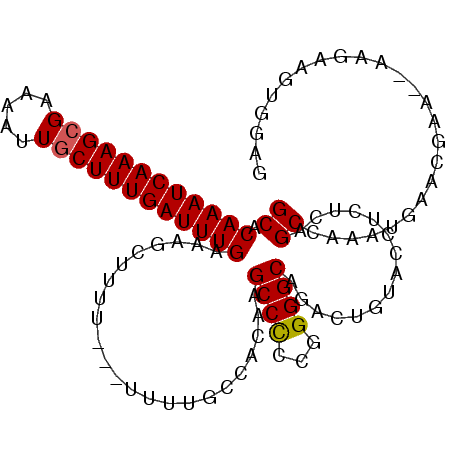

| Location | 18,341,764 – 18,341,870 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 81.35 |
| Mean single sequence MFE | -30.74 |
| Consensus MFE | -24.62 |
| Energy contribution | -25.50 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18341764 106 - 23771897 CUCCAAUUCCU--UUCGUUCAUUUGGCUGAGAGGUACAGUCUGCCCCGGGGGCUUGUGGCAAAAAAAAAAAGCUUUCAAAUCAAAGCAAUUUUCGCUUUGAUUUGUGC ........(((--((((..(....)..)))))))....(((.((((...))))....)))...........((...(((((((((((.......))))))))))).)) ( -30.90) >DroSec_CAF1 126156 93 - 1 CUCCGCUUCUU------------UGGCUGAGAGGUACAGUCUGCCCCGCGGGCUUGUGGCAAAA---AAAAGCUUUCAAAUCAAAGCAAUUUUCGCUUUGAUUUGUGC (((.(((....------------.))).))).(.((((((((((...)))))).)))).)....---....((...(((((((((((.......))))))))))).)) ( -32.30) >DroSim_CAF1 136365 104 - 1 CUCCGCUUCUU--UUCGUUCAUUUCGCUGAGAGGUACAGUCUGCCCCGCGGGCUUGUGGCAAAAA--AAAAGCUUUCAAAUCAAAGCAAUUUUCGCUUUGAUUUGUGC ..(((((((((--..((.......))..)))))))(((((((((...)))))).)))))......--....((...(((((((((((.......))))))))))).)) ( -32.20) >DroEre_CAF1 121785 103 - 1 CUCCACUUCUC--UGCGUUCAUUUGGCUGAGAGGUACAGUCAGCCCCGGGGGCUUGUGGCAACA---AAAAGCUUUCAAAUCAAAGCAAUUUACGCUUUGAUUUGUGC ....(((((((--.((.........)).)))))))...((((((((...))))...))))....---....((...(((((((((((.......))))))))))).)) ( -33.80) >DroYak_CAF1 132508 104 - 1 CUCCACUUCUACUCUCGUUCAUUUGGCUGAAAGGUACAGUCAGCCCCGGGGGCUUGUGGCAA-A---AUAUGCUUUCAAAUCAAAGAAAUUUACGCUUUGAUUUGUGC ..((((.....((((((.....(((((((.......)))))))...))))))...))))...-.---....((...((((((((((.........)))))))))).)) ( -27.60) >DroAna_CAF1 122914 83 - 1 --------------------AUUUGGCUGGCGGGUACAGUCAGCCCUGGAGGCCUU-CGCAU-A---AAAAGCUUUCAAAUCAAAGCAAUUUACGCUUUGAUUUGUGC --------------------....(((((((.......)))))))..((((((...-.....-.---....))))))((((((((((.......)))))))))).... ( -27.62) >consensus CUCCACUUCUU__UUCGUUCAUUUGGCUGAGAGGUACAGUCAGCCCCGGGGGCUUGUGGCAAAA___AAAAGCUUUCAAAUCAAAGCAAUUUACGCUUUGAUUUGUGC ..((((..................(((((.......))))).(((.....)))..))))............((...(((((((((((.......))))))))))).)) (-24.62 = -25.50 + 0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:55 2006