
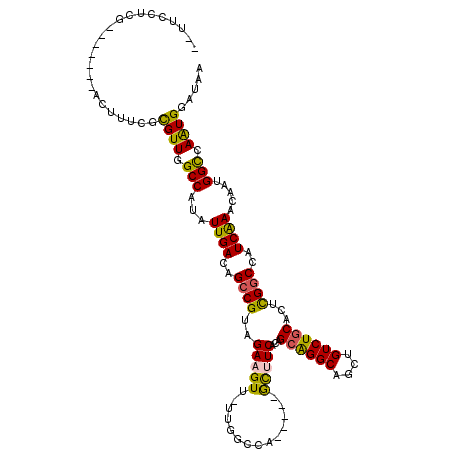
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,339,617 – 18,339,723 |
| Length | 106 |
| Max. P | 0.608791 |

| Location | 18,339,617 – 18,339,723 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.09 |
| Mean single sequence MFE | -35.07 |
| Consensus MFE | -23.58 |
| Energy contribution | -22.81 |
| Covariance contribution | -0.78 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18339617 106 - 23771897 --UUCCCCG-------AUUUUCGCGUUGGCCUUAUUGACAGCCGUAGAAGUUUUUGGCCA-----ACUUCACAGCGGGCAGCUGUCUGCACUCGGCCAUCAAACAAUGGUCAAUGGAUAA --.......-------....((.((((((((...((((..((((..((((((.......)-----)))))...((((((....))))))...))))..)))).....))))))))))... ( -34.50) >DroPse_CAF1 150110 117 - 1 --CACCUCGGCCAGAGCCUUUUUUGUUGGCCAUAUUGACAGCCGUGGAAGUU-UUGGCCAGCACAGCAUCACAGCAGGCAGCUGUCUGCACUCGGCCAUCGAACAAUGGCGAAUGGAUAA --..((..((((.(((.......(((((((((....(((..(....)..)))-.)))))))))..........((((((....)))))).))))))).(((........)))..)).... ( -40.40) >DroEre_CAF1 119714 108 - 1 AUUUCCACA-------AUUUCCGCGUUGGCCUUAUUGACAGCCGUAGAAGUUUUUGGCCA-----GCUUCACAGCAGGCAGCUGUCUGCACUUGGCCAUCAAACAAUGGUCAGUGGAUAA ...(((((.-------........(((((((..(((.((....))...)))....)))))-----))......((((((....))))))...(((((((......))))))))))))... ( -37.40) >DroYak_CAF1 130390 108 - 1 AUUUCCCCG-------AUUUCCGCGUUGGCCUUAUUGACAGGCGUAGAAGUUUUUGGCCA-----GCUUCACAGCAGGCAGCUGUCUCCACUCGGCCAUCAAACAAUGGUCAAUGGAUAA .........-------........(((((((..(((.((....))...)))....)))))-----))...(((((.....))))).((((...((((((......))))))..))))... ( -28.50) >DroAna_CAF1 121078 106 - 1 CCUUCUUUU-------ACC-UCGUGUUGGCCAUAUUGACAGCCGUAGAAGUU-UUGGCCA-----GUUUCACAGCAGGCAGCUGUCUGCACUCGGCCAUCGAACAAUGGCGAAUGAAUAA ..(((.((.-------.((-...((((((((.....(((.((((........-.))))..-----))).....((((((....))))))....)))))....)))..))..)).)))... ( -29.20) >DroPer_CAF1 151135 117 - 1 --CACCUCGGCCAGAGCCUUUUUUGUUGGCCAUAUUGACAGCCGUGGAAGUU-UUGGCCAGCACAGCAUCACAGCAGGCAGCUGUCUGCACUCGGCCAUCGAACAAUGGCGAAUGGAUAA --..((..((((.(((.......(((((((((....(((..(....)..)))-.)))))))))..........((((((....)))))).))))))).(((........)))..)).... ( -40.40) >consensus __UUCCUCG_______ACUUUCGCGUUGGCCAUAUUGACAGCCGUAGAAGUU_UUGGCCA_____GCUUCACAGCAGGCAGCUGUCUGCACUCGGCCAUCAAACAAUGGCCAAUGGAUAA .......................((((.(((...((((..((((..(((((..............)))))...((((((....))))))...))))..)))).....))).))))..... (-23.58 = -22.81 + -0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:51 2006