| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,339,124 – 18,339,218 |
| Length | 94 |
| Max. P | 0.738796 |

| Location | 18,339,124 – 18,339,218 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 88.62 |
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -23.78 |
| Energy contribution | -23.53 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18339124 94 + 23771897 AAGUUAUGGCUACUUGUACAU-GGGCUCGAGGACCGGCAU-CUAAGUGCAGUGA-AAUGCACUCGAGACGAGGGUACAGUUGCCAUUGCAAGGAGUA ..((.(((((.(((.((((.(-.(.((((((((......)-))..(((((....-..)))))))))).).)..))))))).))))).))........ ( -30.90) >DroSec_CAF1 123371 94 + 1 AAGUUAUGGCUACUUGUACAA-GGGCUCGAGGACCGGCAU-CUGAGUGCAGUAA-AAUGCACUCGAGACGACGGUACAGUUGCCAUUGCAAGGAGUA ........(((.((((((...-.(((......((((.(.(-(((((((((....-..))))))).))).).))))......)))..)))))).))). ( -34.40) >DroSim_CAF1 133517 95 + 1 AAGUUAUGGCUACUUGUACAU-GGACUCGAGGACCGGCAUUCUAAGUGCAGUGA-AAUGCACUCGAGACGAGGGUACAGUUGCCAUUGCAAGGAGUA ..((.(((((.(((.((((.(-.(.((((((.....((((.....)))).((..-...)).)))))).).)..))))))).))))).))........ ( -30.50) >DroEre_CAF1 119331 95 + 1 AAGUUAUGGCUACUUGUACAU-GGACUCGAGGACCGCCGU-CUAAGUGCACUGAAAAUGCACUCGAGACGAGGGUACAGUUGCCACUGAAAGGAGUA ......((((.(((.((((..-...(((..((....))((-((.((((((.(....)))))))..))))))).))))))).))))((....)).... ( -30.30) >DroYak_CAF1 129893 95 + 1 AAGUUAUGGCUACUUGUACAU-GGACUCGAGGACCACCAU-CUAAGUGCAGUGAAAAUGCACUCGAGACGAGGGUACAGUUGCCAUUGAAGGCAGUA ..((((((((.(((.((((..-...((((.((....)).(-((.((((((.......))))))..))))))).))))))).)))))....))).... ( -29.00) >DroAna_CAF1 120628 96 + 1 AAGUUAUGGCUACUUGUACAUUGGGCUUGAGGACUGACAG-AUGAGUGCAGUGGAAAUGCACUUGAGACUAGAGUACAGUUGCUUCUCAAAGAAGUA .(((....)))..((((((.((((.(((..(......)..-..(((((((.......)))))))))).)))).)))))).((((((.....)))))) ( -22.50) >consensus AAGUUAUGGCUACUUGUACAU_GGACUCGAGGACCGGCAU_CUAAGUGCAGUGA_AAUGCACUCGAGACGAGGGUACAGUUGCCAUUGAAAGGAGUA ......((((.(((.((((......((((((......).......(((((.......))))))))))......))))))).))))............ (-23.78 = -23.53 + -0.25)



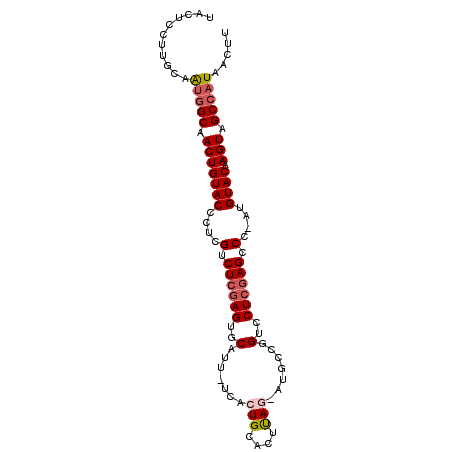
| Location | 18,339,124 – 18,339,218 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 88.62 |
| Mean single sequence MFE | -27.65 |
| Consensus MFE | -21.30 |
| Energy contribution | -21.86 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18339124 94 - 23771897 UACUCCUUGCAAUGGCAACUGUACCCUCGUCUCGAGUGCAUU-UCACUGCACUUAG-AUGCCGGUCCUCGAGCCC-AUGUACAAGUAGCCAUAACUU ...........(((((.(((((((...(((((.(((((((..-....)))))))))-)))..(((......))).-..)))).))).)))))..... ( -28.60) >DroSec_CAF1 123371 94 - 1 UACUCCUUGCAAUGGCAACUGUACCGUCGUCUCGAGUGCAUU-UUACUGCACUCAG-AUGCCGGUCCUCGAGCCC-UUGUACAAGUAGCCAUAACUU ((((...(((((.(((....(.((((.(((((.(((((((..-....)))))))))-))).)))).)....))).-)))))..)))).......... ( -33.90) >DroSim_CAF1 133517 95 - 1 UACUCCUUGCAAUGGCAACUGUACCCUCGUCUCGAGUGCAUU-UCACUGCACUUAGAAUGCCGGUCCUCGAGUCC-AUGUACAAGUAGCCAUAACUU ...........(((((.(((((((.....(((.(((((((..-....)))))))))).....((.(.....).))-..)))).))).)))))..... ( -25.50) >DroEre_CAF1 119331 95 - 1 UACUCCUUUCAGUGGCAACUGUACCCUCGUCUCGAGUGCAUUUUCAGUGCACUUAG-ACGGCGGUCCUCGAGUCC-AUGUACAAGUAGCCAUAACUU ...........(((((.(((((((..((((((.((((((((.....))))))))))-)))).((.(.....).))-..)))).))).)))))..... ( -33.00) >DroYak_CAF1 129893 95 - 1 UACUGCCUUCAAUGGCAACUGUACCCUCGUCUCGAGUGCAUUUUCACUGCACUUAG-AUGGUGGUCCUCGAGUCC-AUGUACAAGUAGCCAUAACUU ...........(((((.(((((((...(((((.(((((((.......)))))))))-)))((((.(.....).))-)))))).))).)))))..... ( -30.80) >DroAna_CAF1 120628 96 - 1 UACUUCUUUGAGAAGCAACUGUACUCUAGUCUCAAGUGCAUUUCCACUGCACUCAU-CUGUCAGUCCUCAAGCCCAAUGUACAAGUAGCCAUAACUU (((((..((((((.....(((.....)))))))))(((((((...((((.((....-..))))))..........)))))))))))).......... ( -14.12) >consensus UACUCCUUGCAAUGGCAACUGUACCCUCGUCUCGAGUGCAUU_UCACUGCACUUAG_AUGCCGGUCCUCGAGCCC_AUGUACAAGUAGCCAUAACUU ...........(((((.(((((((....(.((((((..(.......(((....))).......)..)))))).)....)))).))).)))))..... (-21.30 = -21.86 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:50 2006