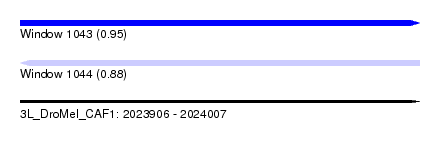
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,023,906 – 2,024,007 |
| Length | 101 |
| Max. P | 0.950366 |

| Location | 2,023,906 – 2,024,007 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 77.28 |
| Mean single sequence MFE | -33.67 |
| Consensus MFE | -20.90 |
| Energy contribution | -21.16 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
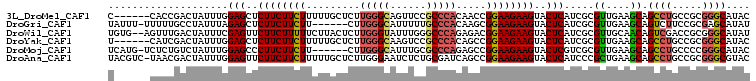
>3L_DroMel_CAF1 2023906 101 + 23771897 C------CACCGACUAUUUGGAGCUCUUCUUCUUUUUGCUCUUGGGCAGUUCCGCCCACAACCGGAAGAAGUACUCAUCGCGUUGAAGCAGCCUGCCGCGGGCAUAC .------...((((......(((..((((((((..(((....(((((......))))))))..))))))))..))).....)))).....(((((...))))).... ( -32.50) >DroGri_CAF1 70842 100 + 1 UAUUU-UUUUUGCCUAUUUAGAGCUCUUCUUCUU------CUUGGGCAUUUUUGCCCACAAGCGGAAGAAGUACUCAUCGCGUUGAAGCAGUCUUCCGCGAGCAUAU .....-....(((.......(((..((((((((.------((((((((....))))))..)).))))))))..))).(((((..((((....))))))))))))... ( -36.90) >DroWil_CAF1 112263 105 + 1 UGUG--AGUUUGACUAUUUCGAGUUCUUCUUUUUCUUACUCUUGGGUAUUUGGGCCCAGAGACGGAAGAAGUACUCAUCGCGUUGCAACAGUCGACCGCGGGCAUAU .(((--.(((.((((.....((((....((((((((..((((.((((......))))))))..)))))))).))))...((...))...))))))))))........ ( -31.40) >DroYak_CAF1 77412 101 + 1 U------CAUCGACUAUUUGGAGCUCUUCUUCUUUUUGCUCUUGGGCAAGUCCGCCCACAGCCGGAAGAAGUACUCAUCGCGUUGAAGCAGCCUGCCGCGGGCAUAC .------..(((((......(((..((((((((....(((..(((((......))))).))).))))))))..))).....)))))....(((((...))))).... ( -35.50) >DroMoj_CAF1 84740 100 + 1 UCAUG-UCUCUGUCUAUUUGGAGCCCUUCUUCUU------CUUGGGCAUUUGCGCCCAGAGCCGGAAGAAGUACUCGUCGCGUUGAAGCAGCCUGCCCCGGGCAUAC .....-..............(((..(((((((((------((.((((......)))))))...))))))))..)))...((......)).(((((...))))).... ( -32.40) >DroAna_CAF1 72827 106 + 1 UACGUC-UAACGACUAUUUGGAGUUCUUCUUCUUUUUGCUCUUGGGAAUCUCUGCGAUCAGCCGGAAGAAGUACUCAUCCCGCUGAAGCAGCCUGCCGCGGGCGUAC ((((((-....))).....((((.....)))).....((((.(((.(....((((..(((((.(((.((.....)).))).))))).))))..).))).))))))). ( -33.30) >consensus U______CAUCGACUAUUUGGAGCUCUUCUUCUUUUUGCUCUUGGGCAUUUCCGCCCACAGCCGGAAGAAGUACUCAUCGCGUUGAAGCAGCCUGCCGCGGGCAUAC ....................(((..((((((((.........(((((......))))).....))))))))..))).....((....)).((((.....)))).... (-20.90 = -21.16 + 0.25)
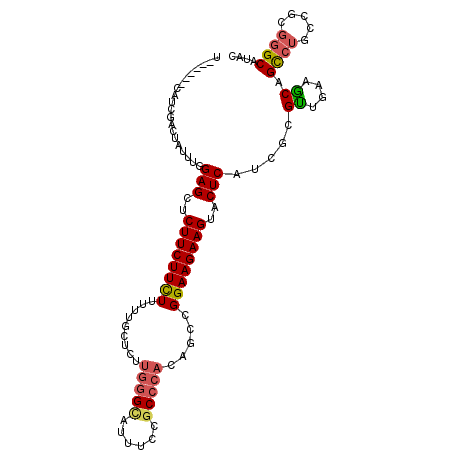

| Location | 2,023,906 – 2,024,007 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.28 |
| Mean single sequence MFE | -32.25 |
| Consensus MFE | -20.20 |
| Energy contribution | -21.01 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
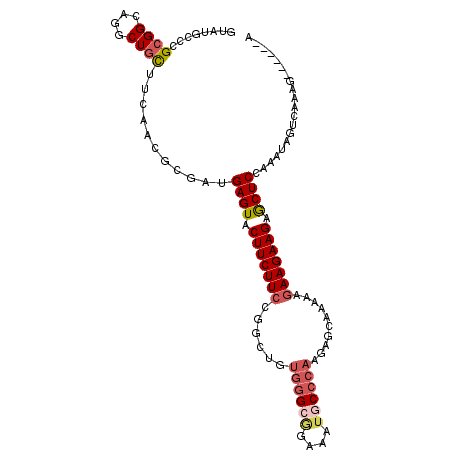
>3L_DroMel_CAF1 2023906 101 - 23771897 GUAUGCCCGCGGCAGGCUGCUUCAACGCGAUGAGUACUUCUUCCGGUUGUGGGCGGAACUGCCCAAGAGCAAAAAGAAGAAGAGCUCCAAAUAGUCGGUG------G ...((((...))))((((((......))...((((.(((((((..(((.((((((....))))))..))).....))))))).)))).....))))....------. ( -33.30) >DroGri_CAF1 70842 100 - 1 AUAUGCUCGCGGAAGACUGCUUCAACGCGAUGAGUACUUCUUCCGCUUGUGGGCAAAAAUGCCCAAG------AAGAAGAAGAGCUCUAAAUAGGCAAAAA-AAAUA ...((((((((((((....))))..))))).((((.(((((((..((..((((((....))))))))------..))))))).)))).......)))....-..... ( -36.20) >DroWil_CAF1 112263 105 - 1 AUAUGCCCGCGGUCGACUGUUGCAACGCGAUGAGUACUUCUUCCGUCUCUGGGCCCAAAUACCCAAGAGUAAGAAAAAGAAGAACUCGAAAUAGUCAAACU--CACA ..............(((((((((...))..(((((.((((((.(..(((((((........))).))))...)...)))))).))))).))))))).....--.... ( -26.40) >DroYak_CAF1 77412 101 - 1 GUAUGCCCGCGGCAGGCUGCUUCAACGCGAUGAGUACUUCUUCCGGCUGUGGGCGGACUUGCCCAAGAGCAAAAAGAAGAAGAGCUCCAAAUAGUCGAUG------A ...((((...))))((((((......))...((((.(((((((..(((.((((((....))))))..))).....))))))).)))).....))))....------. ( -34.60) >DroMoj_CAF1 84740 100 - 1 GUAUGCCCGGGGCAGGCUGCUUCAACGCGACGAGUACUUCUUCCGGCUCUGGGCGCAAAUGCCCAAG------AAGAAGAAGGGCUCCAAAUAGACAGAGA-CAUGA ((.(((..(((((.....)))))...)))))(((..(((((((...((.((((((....))))))))------..)))))))..)))..............-..... ( -31.90) >DroAna_CAF1 72827 106 - 1 GUACGCCCGCGGCAGGCUGCUUCAGCGGGAUGAGUACUUCUUCCGGCUGAUCGCAGAGAUUCCCAAGAGCAAAAAGAAGAAGAACUCCAAAUAGUCGUUA-GACGUA .(((.((((((((.....)))...)))))..((((.(((((((..(((((((.....))))......))).....))))))).))))......(((....-)))))) ( -31.10) >consensus GUAUGCCCGCGGCAGGCUGCUUCAACGCGAUGAGUACUUCUUCCGGCUGUGGGCGGAAAUGCCCAAGAGCAAAAAGAAGAAGAGCUCCAAAUAGUCAAAG______A ........((((....))))...........((((.(((((((......((((((....))))))..........))))))).)))).................... (-20.20 = -21.01 + 0.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:26 2006