
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,325,761 – 18,325,878 |
| Length | 117 |
| Max. P | 0.947104 |

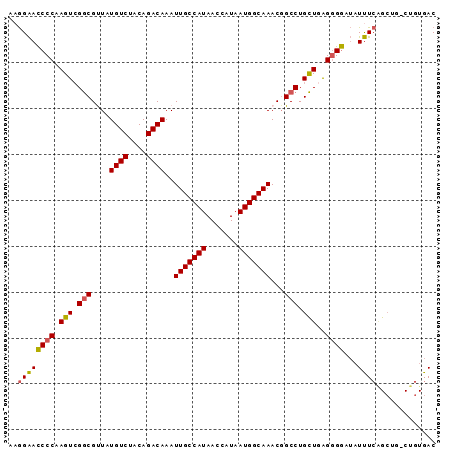
| Location | 18,325,761 – 18,325,853 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 90.97 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -25.14 |
| Energy contribution | -24.95 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18325761 92 - 23771897 AUGGAACCCCAAGUCGGCGUUAUGUCUACAGACAAAUUGCCAUAACCAUAAUGGCAAACUGCCUGCUAAGGGGAUAUUUCAGCUG-CUGUGAC .(((((((((.(((.(((....((((....))))..(((((((.......)))))))...))).)))..))))...)))))....-....... ( -28.20) >DroSec_CAF1 109999 92 - 1 AAGGAACCCCAAGUCGGCGUUAUGUCUACAGACAAAUUGCCAUAACCAUAAUGGCAAACGGCCUGCUGAGGGGAUAUUUCAGCUG-CUGUGAC ..((((((((.(((.(((....((((....))))..(((((((.......)))))))...))).)))..))))...)))).....-....... ( -28.80) >DroSim_CAF1 119847 92 - 1 AAGGAACCCCAAGUCGGCGUUAUGUCUACAGACAAAUUGCCAUAACCAUAAUGGCAAACGGCCUGCUGAGGGGAUAUUUCAGCUG-CUGUGAC ..((((((((.(((.(((....((((....))))..(((((((.......)))))))...))).)))..))))...)))).....-....... ( -28.80) >DroEre_CAF1 105829 91 - 1 AAGGAACCCCAAGUCGGCGCUAUGUCUACAGACAAAUUGCCAUAACCAUAAUGGCAAACGGCCUGCUGAG-GGAUAUUUCAGCUG-CUGUGAC ..((....))..(((.......((((....))))..(((((((.......)))))))(((((..((((((-(....))))))).)-))))))) ( -28.80) >DroYak_CAF1 116336 92 - 1 GAGGAACCCCAAGUCGGCGCUAUGUCUACAGACAAAUUGCCAUAACCAUAAUGGCAAACGGCCUGCUGAGGGGAUAUUUCUGCUG-CUGUGAC .(((((((((.(((.(((....((((....))))..(((((((.......)))))))...))).)))..))))...)))))....-....... ( -29.60) >DroAna_CAF1 107940 91 - 1 CCAGGAUCCCAAGUAGACGUUAUGUCUACAGACAAAUUGCCAUAACCAUAAUGGCAAACGGCCUGUUGGGGGAC--UCUGGGAUGCCUGUGAC ((((..((((..((((((.....)))))).......(((((((.......)))))))....((....)))))).--.))))............ ( -32.30) >consensus AAGGAACCCCAAGUCGGCGUUAUGUCUACAGACAAAUUGCCAUAACCAUAAUGGCAAACGGCCUGCUGAGGGGAUAUUUCAGCUG_CUGUGAC ..((((((((.(((.(((....((((....))))..(((((((.......)))))))...))).)))..))))...))))............. (-25.14 = -24.95 + -0.19)


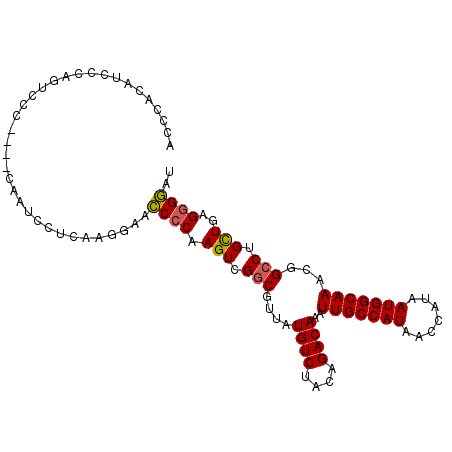
| Location | 18,325,778 – 18,325,878 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 86.58 |
| Mean single sequence MFE | -32.55 |
| Consensus MFE | -24.98 |
| Energy contribution | -24.90 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.94 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18325778 100 - 23771897 ACCCACAUCCCAGUCCC----CAAUCCACAUGGAACCCCAAGUCGGCGUUAUGUCUACAGACAAAUUGCCAUAACCAUAAUGGCAAACUGCCUGCUAAGGGGAU ............(((((----(..(((....)))......(((.(((....((((....))))..(((((((.......)))))))...))).)))..)))))) ( -32.60) >DroSec_CAF1 110016 99 - 1 ACCCAC-UCCCAGUCCC----CAAUCCUCAAGGAACCCCAAGUCGGCGUUAUGUCUACAGACAAAUUGCCAUAACCAUAAUGGCAAACGGCCUGCUGAGGGGAU ......-.....(((((----(..(((....)))......(((.(((....((((....))))..(((((((.......)))))))...))).)))..)))))) ( -32.50) >DroSim_CAF1 119864 100 - 1 ACCCACAUCCCAGUCCC----CAAUCCUCAAGGAACCCCAAGUCGGCGUUAUGUCUACAGACAAAUUGCCAUAACCAUAAUGGCAAACGGCCUGCUGAGGGGAU ............(((((----(..(((....)))......(((.(((....((((....))))..(((((((.......)))))))...))).)))..)))))) ( -32.50) >DroEre_CAF1 105846 99 - 1 ACCCACAUCCCAGCCCC----CAAAACUCAAGGAACCCCAAGUCGGCGCUAUGUCUACAGACAAAUUGCCAUAACCAUAAUGGCAAACGGCCUGCUGAG-GGAU ......(((((......----..........((....))...((((((((.((((....))))..(((((((.......)))))))..)))..))))))-)))) ( -28.20) >DroYak_CAF1 116353 104 - 1 ACCCACAUCCCAGUCCCCCUCCGAUUCUCGAGGAACCCCAAGUCGGCGCUAUGUCUACAGACAAAUUGCCAUAACCAUAAUGGCAAACGGCCUGCUGAGGGGAU .................((((........))))..((((.(((.(((....((((....))))..(((((((.......)))))))...))).)))..)))).. ( -33.00) >DroAna_CAF1 107957 92 - 1 ACCCAAACCCCA-----------AGAGUGCCAGGAUCCCAAGUAGACGUUAUGUCUACAGACAAAUUGCCAUAACCAUAAUGGCAAACGGCCUGUUGGGGGAC- .......(((((-----------(.((.(((..........((((((.....)))))).......(((((((.......)))))))..))))).))))))...- ( -36.50) >consensus ACCCACAUCCCAGUCCC____CAAUCCUCAAGGAACCCCAAGUCGGCGUUAUGUCUACAGACAAAUUGCCAUAACCAUAAUGGCAAACGGCCUGCUGAGGGGAU ...................................((((.(((.(((....((((....))))..(((((((.......)))))))...))).)))..)))).. (-24.98 = -24.90 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:41 2006