
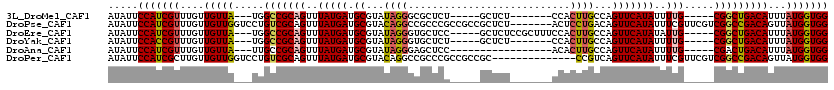
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,321,652 – 18,321,752 |
| Length | 100 |
| Max. P | 0.908096 |

| Location | 18,321,652 – 18,321,752 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.84 |
| Mean single sequence MFE | -34.35 |
| Consensus MFE | -17.74 |
| Energy contribution | -17.55 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
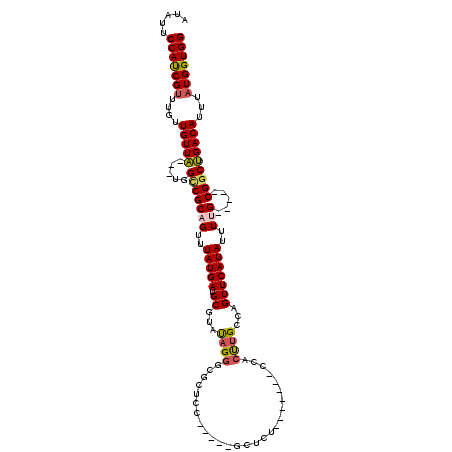
>3L_DroMel_CAF1 18321652 100 - 23771897 AUAUUCCAUCGUUUGUUGUUA---UGGCCGCAGUUUAUGAUGCGUAUAGGGCGCUCU-----GCUCU-------CCACUUGCCAGUUCAUAUUUUG-----CGGCUGACAUUUAUGGUGG .....(((((((....((((.---.((((((((..(((((.(((...(((((.....-----)))))-------.....)))....)))))..)))-----)))))))))...))))))) ( -36.00) >DroPse_CAF1 127862 113 - 1 AUAUUCCAUCGUUUGUUGUUGGUCCUGUCGCAGUUUAUGAUGCGUACAGGCCGCCCGCCGCCGCUCU-------ACUCCUGACAGUUCAUAUUUCGUUCGUCGGCCGACAGUUAUGGUGG .....(((((((((((((..((.(((((((((........)))).)))))))....((((.((..(.-------.....(((....)))......)..)).))))))))))..))))))) ( -35.20) >DroEre_CAF1 101757 107 - 1 AUAUUCCAUCGUUUGUUGUUA---UGGCCGCAGUUUAUGAUGCGUAUAGGGUGCUCC-----GCUCUCCGCUUUCCACUUGCCAGUUCAUAUAUUG-----CGGCUGACAUUUAUGGUGG .....(((((((....((((.---.(((((((((.(((((.(((...((((((...)-----))))).))).....(((....)))))))).))))-----)))))))))...))))))) ( -36.30) >DroYak_CAF1 112245 100 - 1 AUAUUCCACCGUUUGUUGUUA---UGGCCGCAGUUUAUGAUGCGUAUAGGGUGCUCU-----GCUCU-------CCACUUGCCAGUUCAUAUUUUG-----CGGCUGACAUUUAUGGUGG .....(((((((....((((.---.((((((((..(((((.(((...(((((.....-----)))))-------.....)))....)))))..)))-----)))))))))...))))))) ( -37.30) >DroAna_CAF1 100834 95 - 1 AUAUUCCAUCGUUUGUUGUUA---UUGCCGCAGUUUAUGAUGCGUAUAGGGAGCUCC-----------------ACACUUGCCAGUUCAUAUUUUG-----CGACUGACAUUUAUGGUGG .....(((((((....(((((---..(.(((((..(((((.(((.....((....))-----------------.....)))....)))))..)))-----)).))))))...))))))) ( -26.10) >DroPer_CAF1 129321 106 - 1 AUAUUCCAUCGCUUGUUGUUGGUCCUGUCGCAGUUUAUGAUGCGUACAGGCCGCCCGCCGCCGC--------------CCGUCAGUUCAUAUUUCGUUCGUCGGCCGACAGUUAUGGUGG .....((((.(((.((((..((.(((((((((........)))).)))))))....((((.((.--------------.((.............))..)).))))))))))).))))... ( -35.22) >consensus AUAUUCCAUCGUUUGUUGUUA___UGGCCGCAGUUUAUGAUGCGUAUAGGGCGCUCC_____GCUCU_______CCACUUGCCAGUUCAUAUUUUG_____CGGCUGACAUUUAUGGUGG .....(((((((....(((((.....(((((((..(((((.((...((((...........................))))...)))))))..))).....)))))))))...))))))) (-17.74 = -17.55 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:37 2006