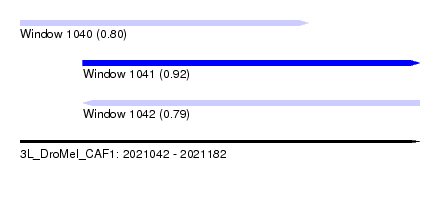
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,021,042 – 2,021,182 |
| Length | 140 |
| Max. P | 0.921891 |

| Location | 2,021,042 – 2,021,143 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.68 |
| Mean single sequence MFE | -21.47 |
| Consensus MFE | -10.75 |
| Energy contribution | -10.78 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.800264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2021042 101 + 23771897 -----------AUUUAGGC-------AUAUACUUACCGCCGUGCGAAAUUUGUUUUUCUGAGCUUUAGAUAGUUUUGACCACAUCUUGGCUGCUCUUUGCAGCCUUUUCUUUACGUCUA- -----------...(((((-------..............((((((((((......(((((...))))).)))))))..))).....((((((.....))))))..........)))))- ( -22.30) >DroSec_CAF1 71476 101 + 1 -----------CUUUAGGC-------AUACACUUACCGCCGUGCGAAAUUUGUUUUUCUGAGCUUUAGAUAGUUUUGACCACAUCUUGGCUGCUCUUUGCAGCCUUUUCUUUACGUCUC- -----------....((((-------...(((........))).((((..(((...((.(((((......))))).))..)))....((((((.....)))))).)))).....)))).- ( -21.70) >DroSim_CAF1 86615 101 + 1 -----------CUUUAGGU-------AUACACUUACCGCCGUGCGAAAUUUGUUUUUCUGAGCUUUAGAUAGUUUUGACCACAUCUUGGCUGCUCUUUGCAGCCUUUUCUUUACGUCUC- -----------.....(((-------(......))))(.((((.((((..(((...((.(((((......))))).))..)))....((((((.....)))))).))))..)))).)..- ( -24.00) >DroEre_CAF1 77976 101 + 1 -----------AUUUAAAC-------ACGCACUUACCAUCAUUCGAAAUUUGCUUUUUUGAGCUUCCGUGAGUUUUGACCACAUCUUGGCUGCUCUUUGCAGCCUUUUCUUUACGUCUA- -----------........-------....((((((.....(((((((.......))))))).....))))))...(((........((((((.....))))))..........)))..- ( -21.27) >DroYak_CAF1 74573 101 + 1 -----------AUUUAAGA-------ACACACUUACCACCUUCCGAAACAUAUUUUUCUGAGUUUUCGAGAGUUUUGACCACAUUCUGGCUGCUCUUUGCAGCCUUUUCUUAACGUCUA- -----------..((((((-------(............((..(((((...((((....)))))))))..))...............((((((.....))))))..)))))))......- ( -21.10) >DroAna_CAF1 69261 118 + 1 UUUCAGAUACGUUUUAGGAGAACAAAAUAACCUUACCUCUUUACGGUAUCGAUCUUGAUCGUCCUUUGAUAGUUUAACCCAUUCUUUGGUACCUUUUUGGAGUGAAA--GCAGCGUCCCC .....(((((((...(((((................))))).)).)))))(((.(((....(((((..(.........(((.....))).......)..))).))..--.))).)))... ( -18.48) >consensus ___________AUUUAGGC_______AUACACUUACCGCCGUGCGAAAUUUGUUUUUCUGAGCUUUAGAUAGUUUUGACCACAUCUUGGCUGCUCUUUGCAGCCUUUUCUUUACGUCUA_ ..........................................(((...........((.(((((......))))).)).........((((((.....)))))).........))).... (-10.75 = -10.78 + 0.03)

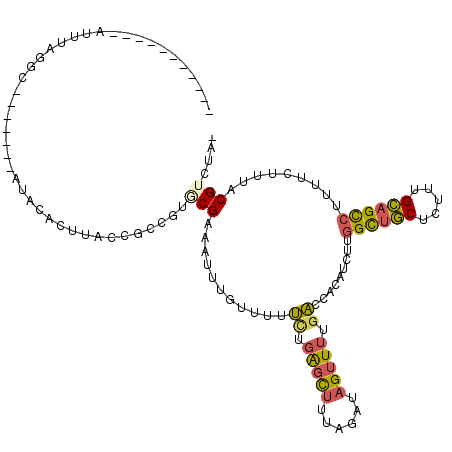
| Location | 2,021,064 – 2,021,182 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.79 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -14.08 |
| Energy contribution | -14.12 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2021064 118 + 23771897 GUGCGAAAUUUGUUUUUCUGAGCUUUAGAUAGUUUUGACCACAUCUUGGCUGCUCUUUGCAGCCUUUUCUUUACGUCUA--CGAUGGUGUUCCUCCUUUGGUACUCGCGCAAAAAUCCAA ((((((...........(.(((((......))))).)((((......((((((.....)))))).........((....--)).))))((.((......)).)))))))).......... ( -29.50) >DroSec_CAF1 71498 118 + 1 GUGCGAAAUUUGUUUUUCUGAGCUUUAGAUAGUUUUGACCACAUCUUGGCUGCUCUUUGCAGCCUUUUCUUUACGUCUC--CGGUGGCGUUCCUCCUUUGGAACUCUCGCAAAAAUCUUA .(((((..........((.(((((......))))).))((((.....((((((.....)))))).........((....--)))))).(((((......)))))..)))))......... ( -33.60) >DroSim_CAF1 86637 118 + 1 GUGCGAAAUUUGUUUUUCUGAGCUUUAGAUAGUUUUGACCACAUCUUGGCUGCUCUUUGCAGCCUUUUCUUUACGUCUC--CGGUGGCGUUCCUCCUUUGGAACUCUCGCAAAAAUCUUA .(((((..........((.(((((......))))).))((((.....((((((.....)))))).........((....--)))))).(((((......)))))..)))))......... ( -33.60) >DroEre_CAF1 77998 118 + 1 AUUCGAAAUUUGCUUUUUUGAGCUUCCGUGAGUUUUGACCACAUCUUGGCUGCUCUUUGCAGCCUUUUCUUUACGUCUA--AGCUGGCGUUUCUUCUAUGGUACUCGCGCAGAAAUCCUA .(((.......((((....))))...(((((((....((((......((((((.....))))))........(((((..--....)))))........)))))))))))..)))...... ( -33.70) >DroYak_CAF1 74595 118 + 1 UUCCGAAACAUAUUUUUCUGAGUUUUCGAGAGUUUUGACCACAUUCUGGCUGCUCUUUGCAGCCUUUUCUUAACGUCUA--GGUUGGCGUUUCUUCUAUGGUACUCACGCAAAAAUCCUA (((.((((......)))).)))((((((.((((....((((......((((((.....)))))).......((((((..--....)))))).......)))))))).)).))))...... ( -28.70) >DroAna_CAF1 69301 118 + 1 UUACGGUAUCGAUCUUGAUCGUCCUUUGAUAGUUUAACCCAUUCUUUGGUACCUUUUUGGAGUGAAA--GCAGCGUCCCCAUAUUUGAAUUAUUUCUUCGGAACUCACGCAUGAAACCAA ....(((((((((....)))(((....))).................))))))...((((.((....--)).((((...((....))......(((....)))...))))......)))) ( -20.40) >consensus GUGCGAAAUUUGUUUUUCUGAGCUUUAGAUAGUUUUGACCACAUCUUGGCUGCUCUUUGCAGCCUUUUCUUUACGUCUA__CGGUGGCGUUCCUCCUUUGGAACUCACGCAAAAAUCCUA ................((.(((((......))))).))((.......((((((.....))))))........(((((........))))).........))................... (-14.08 = -14.12 + 0.03)
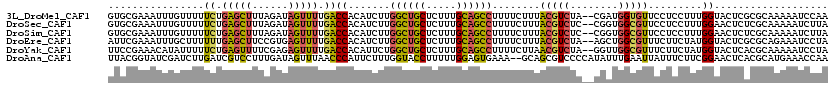
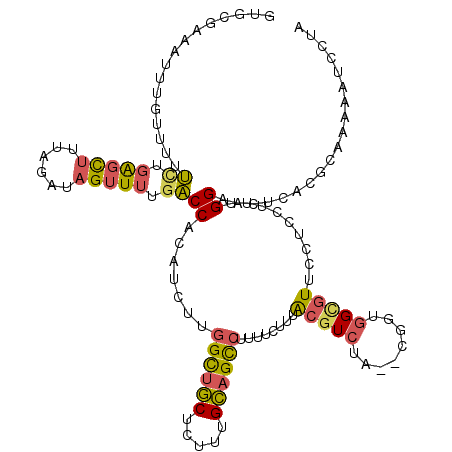
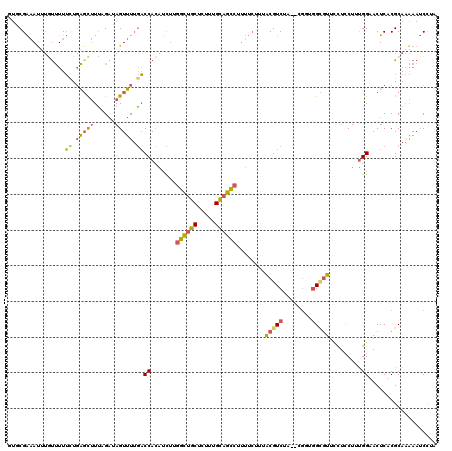
| Location | 2,021,064 – 2,021,182 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.79 |
| Mean single sequence MFE | -29.91 |
| Consensus MFE | -11.81 |
| Energy contribution | -11.78 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2021064 118 - 23771897 UUGGAUUUUUGCGCGAGUACCAAAGGAGGAACACCAUCG--UAGACGUAAAGAAAAGGCUGCAAAGAGCAGCCAAGAUGUGGUCAAAACUAUCUAAAGCUCAGAAAAACAAAUUUCGCAC (((..((((((.((..((.((......)).))(((((((--....)).........((((((.....)))))).....)))))..............)).))))))..)))......... ( -27.70) >DroSec_CAF1 71498 118 - 1 UAAGAUUUUUGCGAGAGUUCCAAAGGAGGAACGCCACCG--GAGACGUAAAGAAAAGGCUGCAAAGAGCAGCCAAGAUGUGGUCAAAACUAUCUAAAGCUCAGAAAAACAAAUUUCGCAC .........((((((((((((......)))))(((((((--....)).........((((((.....)))))).....))))).............................))))))). ( -36.60) >DroSim_CAF1 86637 118 - 1 UAAGAUUUUUGCGAGAGUUCCAAAGGAGGAACGCCACCG--GAGACGUAAAGAAAAGGCUGCAAAGAGCAGCCAAGAUGUGGUCAAAACUAUCUAAAGCUCAGAAAAACAAAUUUCGCAC .........((((((((((((......)))))(((((((--....)).........((((((.....)))))).....))))).............................))))))). ( -36.60) >DroEre_CAF1 77998 118 - 1 UAGGAUUUCUGCGCGAGUACCAUAGAAGAAACGCCAGCU--UAGACGUAAAGAAAAGGCUGCAAAGAGCAGCCAAGAUGUGGUCAAAACUCACGGAAGCUCAAAAAAGCAAAUUUCGAAU ..(..((((((...((((((((((......(((.(....--..).)))........((((((.....))))))....))))))....)))).))))))..)................... ( -28.50) >DroYak_CAF1 74595 118 - 1 UAGGAUUUUUGCGUGAGUACCAUAGAAGAAACGCCAACC--UAGACGUUAAGAAAAGGCUGCAAAGAGCAGCCAGAAUGUGGUCAAAACUCUCGAAAACUCAGAAAAAUAUGUUUCGGAA ...((.((((.((.((((((((((.....((((.(....--..).)))).......((((((.....))))))....))))))....)))).)))))).)).((((......)))).... ( -29.30) >DroAna_CAF1 69301 118 - 1 UUGGUUUCAUGCGUGAGUUCCGAAGAAAUAAUUCAAAUAUGGGGACGCUGC--UUUCACUCCAAAAAGGUACCAAAGAAUGGGUUAAACUAUCAAAGGACGAUCAAGAUCGAUACCGUAA ((((......(((.(.(((((....................))))).))))--.......))))...((((((...((.(((......)))))...)).((((....)))).)))).... ( -20.77) >consensus UAGGAUUUUUGCGAGAGUACCAAAGAAGGAACGCCAACG__GAGACGUAAAGAAAAGGCUGCAAAGAGCAGCCAAGAUGUGGUCAAAACUAUCUAAAGCUCAGAAAAACAAAUUUCGCAC ...((((...((....))......................................((((((.....)))))).......)))).................................... (-11.81 = -11.78 + -0.03)

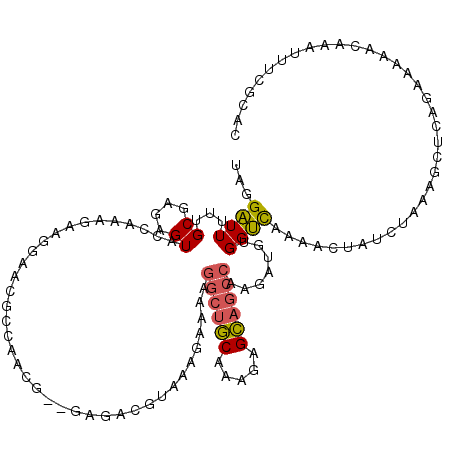
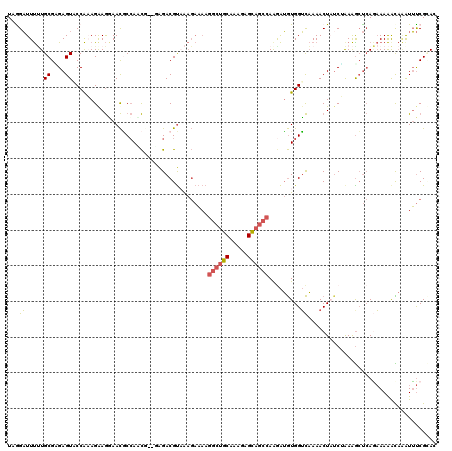
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:24 2006