| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,308,656 – 18,308,748 |
| Length | 92 |
| Max. P | 0.956051 |

| Location | 18,308,656 – 18,308,748 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.66 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -26.58 |
| Energy contribution | -26.22 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -4.30 |
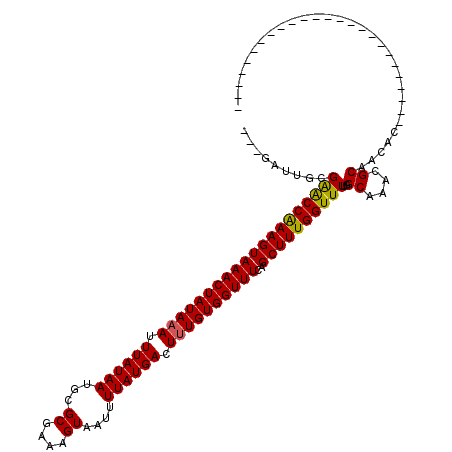
| Structure conservation index | 0.85 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18308656 92 + 23771897 ---AAUUCCGAGCCAAAGUAAACUAUAAAUUUAUAAUGCGCGAAAGUAAUUUUAUGACUUUGUGGUUUCAGCUUUGGUUUUGGCAAACGCAACAU------------------------- ---....((((((((((((((((((((((.((((((...((....))....)))))).))))))))))..))))))).)))))............------------------------- ( -27.70) >DroPse_CAF1 112188 119 + 1 CCCGAUUAUGAACCAAAGUAAACUAUAAAUUUAUAAUGCGCGAAAGUAAUUUUAUGACUUUGUGGUUUUAGCUUUGGUUUUGGCACA-GCUAUACGUUUCUAUGUAUCUGUGUAUCUAUG ...(((...((((((((((((((((((((.((((((...((....))....)))))).))))))))))..))))))))))..(((((-(..((((((....))))))))))))))).... ( -37.20) >DroSec_CAF1 92473 92 + 1 ---UAUUGCGAGCCAAAGUAAACUAUAAAUUUAUAAUGCGCGAAAGUAAUUUUAUGACUUUGUGGUUUCAGCUUUGGUUUUGGCAAACGCAACAC------------------------- ---..((((((((((((((((((((((((.((((((...((....))....)))))).))))))))))..))))))))))..)))).........------------------------- ( -30.00) >DroSim_CAF1 101878 92 + 1 ---UAUUGCGAGCCAAAGUAAACUAUAAAUUUAUAAUGCGCGAAAGUAAUUUUAUGACUAUGUGGUUUCAGCUUUGGUUUUGGCAAACGCAACAC------------------------- ---..((((((((((((((((((((((.(.((((((...((....))....)))))).).))))))))..))))))))))..)))).........------------------------- ( -26.50) >DroAna_CAF1 89368 87 + 1 ----AUUGUGGACCGAAGUAAACUAUAAAUUUAUAAUGCGCGAAAGUAAUUUUAUGACUUUGUGGUUUUAGCUUUGGUUUUGGCAAACGCC----------------------------- ----.....((((((((((((((((((((.((((((...((....))....)))))).))))))))))..)))))))))).(((....)))----------------------------- ( -28.00) >DroPer_CAF1 113266 119 + 1 CCCGAUUAUGAACCAAAGUAAACUAUAAAUUUAUAAUGCGCGAAAGUAAUUUUAUGACUUUGUGGUUUUAGCUUUGGUUUUGGCACA-GCUAUACGUUUCUAUGUAUCUGUGUAUCUGUG ...(((...((((((((((((((((((((.((((((...((....))....)))))).))))))))))..))))))))))..(((((-(..((((((....))))))))))))))).... ( -37.20) >consensus ___GAUUGCGAACCAAAGUAAACUAUAAAUUUAUAAUGCGCGAAAGUAAUUUUAUGACUUUGUGGUUUCAGCUUUGGUUUUGGCAAACGCAACAC_________________________ .........((((((((((((((((((((.((((((...((....))....)))))).))))))))))..))))))))))..((....)).............................. (-26.58 = -26.22 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:34 2006