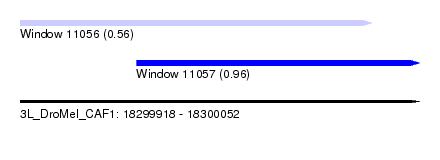
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,299,918 – 18,300,052 |
| Length | 134 |
| Max. P | 0.963243 |

| Location | 18,299,918 – 18,300,036 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.38 |
| Mean single sequence MFE | -25.87 |
| Consensus MFE | -22.78 |
| Energy contribution | -22.67 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
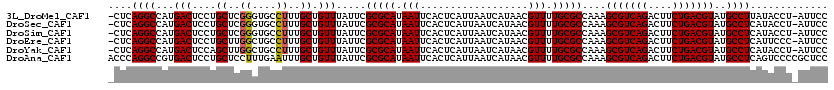
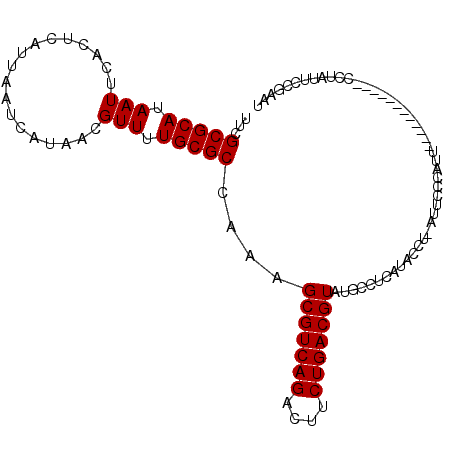
>3L_DroMel_CAF1 18299918 118 + 23771897 -CUCAGGCCAUGACUCCUGCUCGGGUGCCUUUGCUGUUUAUUCGCGCAUAAUUCACUCAUUAAUCAUAACGUUUUGCGCCAAAGCGUCAGACUUCUGACGUAUGCCUUAUACCU-AUUCC -...((((....((....(((..(((((...(((((......)).)))((((......)))).............)))))..)))(((((....)))))))..)))).......-..... ( -26.10) >DroSec_CAF1 83692 118 + 1 -CUCAGGCCAUGACUCCUGCUCGGGUGCCUUUGCUGUUUAUUCGCGCAUAAUUCACUCAUUAAUCAUAACGUUUUGCGCCAAAGCGUCAGACUUCUGACGUAUGCCUCAUACCU-AUUCC -...((((....((....(((..(((((...(((((......)).)))((((......)))).............)))))..)))(((((....)))))))..)))).......-..... ( -26.10) >DroSim_CAF1 93075 118 + 1 -CUCAGGCCAUGACUCCUGCUCGGGUGCCUUUGCUGUUUAUUCGCGCAUAAUUCACUCAUUAAUCAUAACGUUUUGCGCCAAAGCGUCAGACUUCUGACGUAUGCCUCAUACCU-AUUCC -...((((....((....(((..(((((...(((((......)).)))((((......)))).............)))))..)))(((((....)))))))..)))).......-..... ( -26.10) >DroEre_CAF1 80555 118 + 1 -CUCAGGCCAUGACUCCUGCUUGGCUGCCUUUGCUGUUUAUUCGCGCAUAAUUCACUCAUUAAUCAUAACGUUUUGCGCCAAAGCGUCAGACUUCUGACGUAUGCCUCAUUCCC-AUUCC -...((((....((....(((((((.......)))........(((((.(((..................))).)))))..))))(((((....)))))))..)))).......-..... ( -24.67) >DroYak_CAF1 89984 118 + 1 -CUCAGGCCAUGACUCCAGCUUGGCUGCCUUUGCUGUUUAUUCGCGCAUAAUUCACUCAUUAAUCAUAACGUUUUGCGCCAAAGCGUCAGACUUCUGACGUAUGCCUCAUACCU-AUUCC -...((((........((((..((....))..)))).......(((((.(((..................))).)))))....(((((((....)))))))..)))).......-..... ( -26.77) >DroAna_CAF1 81355 120 + 1 ACCCAGGCCGUGACUCCUGCUCCUUUGAAUUUGCUGUUUAUUCGCGCAUAAUUCACUCAUUAAUCAUAACGUUUUGCGCCAAAGCGUCAGACUUCUGACGUAUGCCUCAGUCCCCGCUCC .....(((.(.((((...((......((((.........))))(((((.(((..................))).)))))....(((((((....)))))))..))...)))).).))).. ( -25.47) >consensus _CUCAGGCCAUGACUCCUGCUCGGGUGCCUUUGCUGUUUAUUCGCGCAUAAUUCACUCAUUAAUCAUAACGUUUUGCGCCAAAGCGUCAGACUUCUGACGUAUGCCUCAUACCU_AUUCC ....((((...(((....((..((....))..)).))).....(((((.(((..................))).)))))....(((((((....)))))))..))))............. (-22.78 = -22.67 + -0.11)
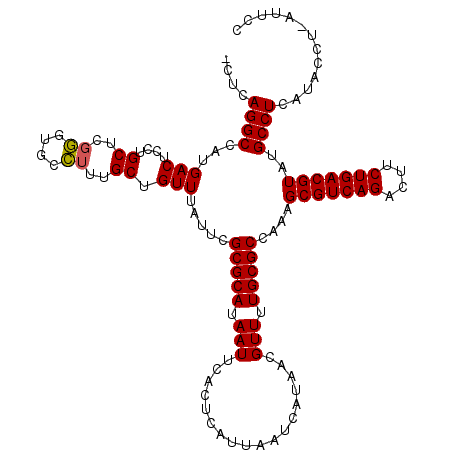
| Location | 18,299,957 – 18,300,052 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 80.41 |
| Mean single sequence MFE | -18.71 |
| Consensus MFE | -16.87 |
| Energy contribution | -16.87 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18299957 95 + 23771897 UUCGCGCAUAAUUCACUCAUUAAUCAUAACGUUUUGCGCCAAAGCGUCAGACUUCUGACGUAUGCCUUAUACCU-AUUCCCAUU------------CCUAUUCCGAAU ((((((((.(((..................))).)))))....(((((((....))))))).............-.........------------........))). ( -17.67) >DroSec_CAF1 83731 95 + 1 UUCGCGCAUAAUUCACUCAUUAAUCAUAACGUUUUGCGCCAAAGCGUCAGACUUCUGACGUAUGCCUCAUACCU-AUUCCCAUU------------CCUAUUCCGAAU ((((((((.(((..................))).)))))....(((((((....))))))).............-.........------------........))). ( -17.67) >DroSim_CAF1 93114 94 + 1 UUCGCGCAUAAUUCACUCAUUAAUCAUAACGUUUUGCGCCAAAGCGUCAGACUUCUGACGUAUGCCUCAUACCU-AUUCCCAUU-------------CUAUUCCGAAU ((((((((.(((..................))).)))))....(((((((....))))))).............-.........-------------.......))). ( -17.67) >DroYak_CAF1 90023 107 + 1 UUCGCGCAUAAUUCACUCAUUAAUCAUAACGUUUUGCGCCAAAGCGUCAGACUUCUGACGUAUGCCUCAUACCU-AUUCCCAUUCCUAUAACCAUUCCUAUUCCGAAU ((((((((.(((..................))).)))))....(((((((....))))))).............-.............................))). ( -17.67) >DroAna_CAF1 81395 101 + 1 UUCGCGCAUAAUUCACUCAUUAAUCAUAACGUUUUGCGCCAAAGCGUCAGACUUCUGACGUAUGCCUCAGUCCCCGCUCC-------ACAUCCAUUUCUAUUCGGGGU ...(((((.(((..................))).)))))....(((((((....)))))))..........(((((....-------...............))))). ( -23.18) >DroPer_CAF1 102539 73 + 1 UUCGCGCAUAAUUCACUCAUUAAUCAUUACGUUUUGCGCCAAAGCGUCAGACUUCUGACGUGUGCCGCCAUGG----------------------------------- ...((((((.....................((((((...))))))(((((....)))))))))))........----------------------------------- ( -18.40) >consensus UUCGCGCAUAAUUCACUCAUUAAUCAUAACGUUUUGCGCCAAAGCGUCAGACUUCUGACGUAUGCCUCAUACCU_AUUCCCAUU____________CCUAUUCCGAAU ...(((((.(((..................))).)))))....(((((((....)))))))............................................... (-16.87 = -16.87 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:26 2006