
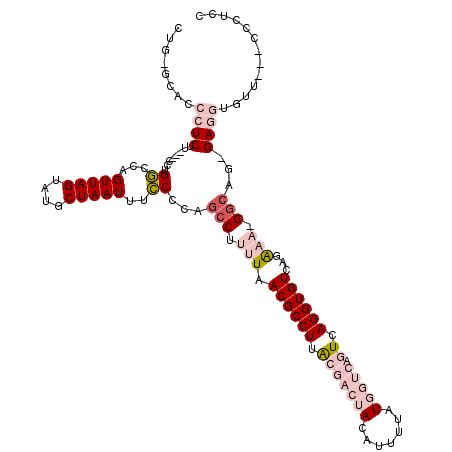
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,295,646 – 18,295,757 |
| Length | 111 |
| Max. P | 0.623096 |

| Location | 18,295,646 – 18,295,757 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.72 |
| Mean single sequence MFE | -38.72 |
| Consensus MFE | -15.56 |
| Energy contribution | -18.12 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
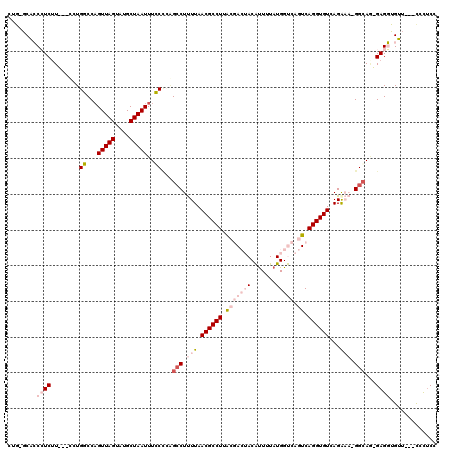
>3L_DroMel_CAF1 18295646 111 - 23771897 CUG-GCACCCUCUU---UCUGUCCAGUUAGUAUGCUAAUUUGCUCAGGCUUUUAACGCCUUACGACUACAUUUUAUGGUCAGUCAGGUGUCAGAAA-GGCAU-GAGGUGUU---CCCUCC ..(-(.((((((..---.(((..(((((((....)))).)))..)))((((((.((((((.(((((((.......))))).)).))))))...)))-)))..-)))).)).---)).... ( -34.80) >DroPse_CAF1 95720 114 - 1 CUG-GCCGCGUCUUGGCCGUGGCCAGUUAGUAUGCUAAUUUCCCGGGCCUCUUAACGCCUAAGUA--ACGUUUUAUGGCCAGUCAGGUGUCAGA---GGCAGCGAGCCGGUUGCUUUUCC (((-((((((((((((((.((((.((((((....)))))).....((((....((((........--.))))....)))).)))).).)))).)---))).))).))))).......... ( -40.10) >DroEre_CAF1 76553 111 - 1 CUG-GCACCCUCUU---UCUGUCCAGUUAGUAUGCUAAUUUGCCCAGCCUUUUAACGCCUUACGACUACAUUUUAUGGUCAGUCAGGUGUCAGGAA-GGCAA-GAGGUGUU---CCCUCC ..(-(.((((((((---..((..(((((((....)))).)))..))(((((((.((((((.(((((((.......))))).)).))))))..))))-)))))-)))).)).---)).... ( -36.80) >DroYak_CAF1 85801 112 - 1 CUGGGCACCCUCUU---ACUGUCCAGUUAGUAUGCUAAUUUGCCCAGCCUUUUAACGCCUUGCGACUACAUUUUAUGGUCAGUCAGGUGUCAGAAA-GGCAG-GAGGUGUU---CCCUCC .((((((......(---((((......)))))........))))))(((((((.((((((.(((((((.......))))).)).))))))..))))-))).(-((((....---.))))) ( -42.04) >DroAna_CAF1 77591 107 - 1 -----CACCCUCGU---CUUGGCCUGUUAGUAUGCUAAUUUCCACUGCCUUUUAACGCCUUACGACUACAUUUUAUGGUCAGUCAGGUGUCAGAAAAGGGAA-GAGG-GUG---CACCCC -----(((((((.(---(.(((...(((((....)))))..)))...((((((.((((((.(((((((.......))))).)).))))))...)))))))).-))))-)))---...... ( -38.40) >DroPer_CAF1 96899 114 - 1 CUG-GCCGCGUCUUGGCCGUGGCCAGUUAGUAUGCUAAUUUCCCGGGCCUCUUAACGCCUAAGUA--ACGCUUUAUGGCCAGUCAGGUGUCAGA---GGCAGUGAGCCGGUUGCUUUUCC (((-(((((.......(((.((..((((((....)))))).)))))((((((..((((((.....--..(((....))).....)))))).)))---))).))).))))).......... ( -40.20) >consensus CUG_GCACCCUCUU___CCUGGCCAGUUAGUAUGCUAAUUUCCCCAGCCUUUUAACGCCUUACGACUACAUUUUAUGGUCAGUCAGGUGUCAGAAA_GGCAG_GAGGUGUU___CCCUCC ........((((........((...(((((....)))))..))...(((.(((.((((((.(((((((.......))))).)).))))))...))).)))...))))............. (-15.56 = -18.12 + 2.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:24 2006