
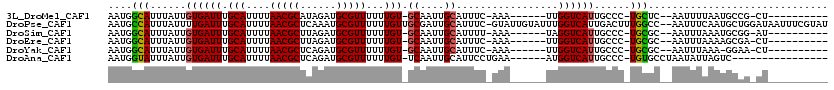
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,293,039 – 18,293,137 |
| Length | 98 |
| Max. P | 0.686804 |

| Location | 18,293,039 – 18,293,137 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.01 |
| Mean single sequence MFE | -24.41 |
| Consensus MFE | -13.34 |
| Energy contribution | -13.15 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
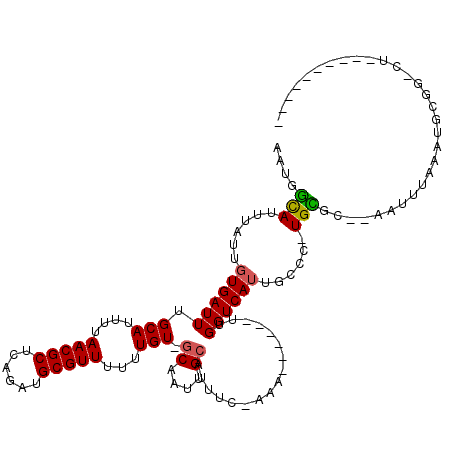
>3L_DroMel_CAF1 18293039 98 - 23771897 AAUGGCAUUUAUUGUGAUUUGCAUUUUAACGCAUAGAUGCGUUUUUUGU-GCAAUUGCAUUUC-AAA------UUGGUCAUUGCCC-UGCUC--AAUUUUAAUGCCG-CU---------- ...((((((...((..(((.((((...((((((....))))))....))-)))))..))....-(((------(((..((......-))..)--))))).)))))).-..---------- ( -27.50) >DroPse_CAF1 92154 117 - 1 AAUGCCAUUUAUUUUGAUUUGCAUUUUAACGCUCAAAUGCGUUUUUUGUUGCGAUUGCAUUUC-GUAUUGUAUUUGGUCAUUGACUUUGGCC--AAUUUCAAUGCUGGAUAAUUUCGUAU ....(((.......((..(((((....(((((......)))))......)))))...))....-((((((...(((((((.......)))))--))...)))))))))............ ( -29.10) >DroSim_CAF1 84936 98 - 1 AAUGGCAUUUAUUGUGAUUUGCAUUUUAACGCUUAGAUGCGUUUUUUGU-GCAAUUGCAUUUU-AAA------UAGGUCAUUGCCC-UGCGC--AAUUUAAAUGCGG-AU---------- (((((((((((..(((..((((((...(((((......)))))....))-))))...)))..)-)))------)..))))))....-..(((--(.......)))).-..---------- ( -27.20) >DroEre_CAF1 73836 98 - 1 AAUGGCAUUUAUUGUGAUUUGCAUUUUAACGCUUAGAUGCGUUUUUUGU-GCAAUUGCAUUUC-AAA------UUGGUCAUUGCCC-UGCGC--AAUUUAAAAGCGA-CU---------- ((((((......((..(((.((((...(((((......)))))....))-)))))..))....-...------...))))))....-..(((--.........))).-..---------- ( -22.59) >DroYak_CAF1 83067 97 - 1 AAUGGCAUUUAUUGUGAUUUGCAUUUUAACGCUCAGAUGCGUUUUUUGU-GCAAUUGCAUUUC-AAA------UUGGUCAUUGCCC-UGCGC--AAUUUAAA-GGAA-CU---------- ((((((......((..(((.((((...(((((......)))))....))-)))))..))....-...------...))))))..((-(....--.......)-))..-..---------- ( -21.99) >DroAna_CAF1 75032 96 - 1 AAUGGUAUUUAUUGUGAUUUGCAUUUUAACGCUCAGAUGCGUUUUUUGU-UCAAUUGCAUUCCUGAA------AUGGUCAUUGCCC-UGUGCCUAAUAUUAGUC---------------- ...(((((....((..(((.(((....(((((......)))))...)))-..)))..))........------..(((....))).-.)))))...........---------------- ( -18.10) >consensus AAUGGCAUUUAUUGUGAUUUGCAUUUUAACGCUCAGAUGCGUUUUUUGU_GCAAUUGCAUUUC_AAA______UUGGUCAUUGCCC_UGCGC__AAUUUAAAUGCGG_CU__________ ....(((......((((((.(((....(((((......)))))...))).((....)).................))))))......))).............................. (-13.34 = -13.15 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:22 2006