
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,279,395 – 18,279,490 |
| Length | 95 |
| Max. P | 0.889231 |

| Location | 18,279,395 – 18,279,490 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.74 |
| Mean single sequence MFE | -19.82 |
| Consensus MFE | -14.60 |
| Energy contribution | -14.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
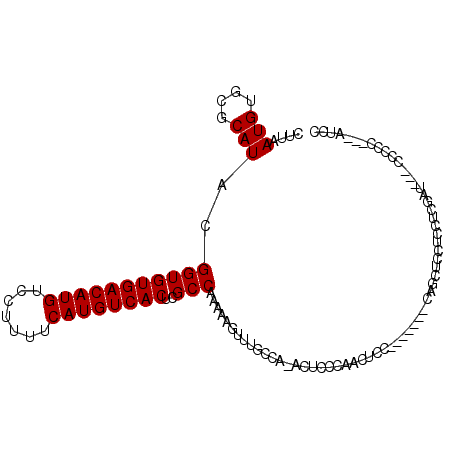
>3L_DroMel_CAF1 18279395 95 + 23771897 CUUAAUGUGCGCAUACGGUGUGACAUGUCCUUUUCAUGUCACUCGCCAAAAAGUUUGC-A-ACUCCCAACUCC-------CAGCUUCUCCUCGAU---CCUCC---AUCC ..........(((.(((((((((((((.......))))))))..))).....)).)))-.-............-------...............---.....---.... ( -16.50) >DroPse_CAF1 73310 103 + 1 CUUAAUGUGCGCAUACGGUGUGACAUGUCCUUUUCAUGUCACUCGCCGAAAAAGUUGCCAAACUCCCGACUGGCCCC---CCCCCCCACCCCGACUGCCCCCC---GUC- ..........(((..(((.((((((((.......))))))))..((((....((((....))))......))))...---..........)))..))).....---...- ( -22.50) >DroEre_CAF1 60690 96 + 1 CUUAAUGUGCGCAUACGGUGUGACAUGUCCUUUUCAUGUCACUCGCCAAAAAGUUUGCCA-ACUCCCAGCUCC-------CAGCUCCUCCUCGAU---CCCCC---AUCC .......((.(((.(((((((((((((.......))))))))..))).....)).)))))-......(((...-------..)))..........---.....---.... ( -18.10) >DroYak_CAF1 69561 103 + 1 CUUAAUGUGCGCAUACGGUGUGACAUGUCCUUUUCAUGUCACUCGCCAAAAAGUUUGCCA-ACUCCCAACUCCCCGCUCACAGCUCCUCCUCGAU---CUCCC---AUCC .....((((.((....(((((((((((.......))))))))..)))....((((....)-)))...........)).)))).............---.....---.... ( -21.70) >DroAna_CAF1 63185 82 + 1 CUUAAUGUGCGCAUACGGUGUGACAUGUCCUUUUCAUGUCACUCGCCAAAAAGUUUCCCA-ACUC------------------------CACGAU---CCUCCUGCGUCC ........(((((...(((((((((((.......))))))))..)))....((((....)-))).------------------------......---.....))))).. ( -19.70) >DroPer_CAF1 74919 92 + 1 CUUAAUGUGCGCAUACGGUGUGACAUGUCCUUUUCAUGUCACUCGCCGAAAAAGUUGCCAAACUCCCGACUGCCC--------------CCCGACUGCCCCCC---GUC- ..........((...((((((((((((.......))))))))..))))....(((((.........)))))))..--------------...(((........---)))- ( -20.40) >consensus CUUAAUGUGCGCAUACGGUGUGACAUGUCCUUUUCAUGUCACUCGCCAAAAAGUUUGCCA_ACUCCCAACUCC_______CAGCUCCUCCUCGAU___CCCCC___AUCC ....(((....)))..(((((((((((.......))))))))..)))............................................................... (-14.60 = -14.60 + -0.00)

| Location | 18,279,395 – 18,279,490 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.74 |
| Mean single sequence MFE | -30.33 |
| Consensus MFE | -15.61 |
| Energy contribution | -16.00 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
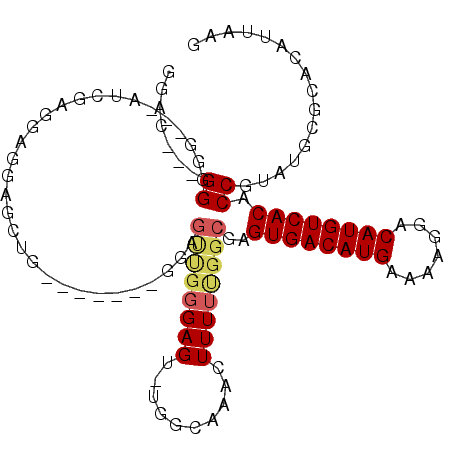
>3L_DroMel_CAF1 18279395 95 - 23771897 GGAU---GGAGG---AUCGAGGAGAAGCUG-------GGAGUUGGGAGU-U-GCAAACUUUUUGGCGAGUGACAUGAAAAGGACAUGUCACACCGUAUGCGCACAUUAAG .(((---(..(.---((...((....((..-------((((((......-.-...))).)))..))..((((((((.......)))))))).))..)).)...))))... ( -23.70) >DroPse_CAF1 73310 103 - 1 -GAC---GGGGGGCAGUCGGGGUGGGGGGG---GGGGCCAGUCGGGAGUUUGGCAACUUUUUCGGCGAGUGACAUGAAAAGGACAUGUCACACCGUAUGCGCACAUUAAG -...---...(.(((..(((.((.(..(((---(..(((((.(....).)))))..))))..).))..((((((((.......)))))))).)))..))).)........ ( -41.20) >DroEre_CAF1 60690 96 - 1 GGAU---GGGGG---AUCGAGGAGGAGCUG-------GGAGCUGGGAGU-UGGCAAACUUUUUGGCGAGUGACAUGAAAAGGACAUGUCACACCGUAUGCGCACAUUAAG (.((---(.((.---.((.((......)).-------)).((..(((((-(....))).)))..))..((((((((.......)))))))).)).))).).......... ( -26.20) >DroYak_CAF1 69561 103 - 1 GGAU---GGGAG---AUCGAGGAGGAGCUGUGAGCGGGGAGUUGGGAGU-UGGCAAACUUUUUGGCGAGUGACAUGAAAAGGACAUGUCACACCGUAUGCGCACAUUAAG .(((---(....---.((......))((.(((.((((...((..(((((-(....))).)))..))..((((((((.......)))))))).)))).))))).))))... ( -28.90) >DroAna_CAF1 63185 82 - 1 GGACGCAGGAGG---AUCGUG------------------------GAGU-UGGGAAACUUUUUGGCGAGUGACAUGAAAAGGACAUGUCACACCGUAUGCGCACAUUAAG (..((((...((---.((((.------------------------(((.-.((....)).))).))))((((((((.......)))))))).))...))))..)...... ( -30.90) >DroPer_CAF1 74919 92 - 1 -GAC---GGGGGGCAGUCGGG--------------GGGCAGUCGGGAGUUUGGCAACUUUUUCGGCGAGUGACAUGAAAAGGACAUGUCACACCGUAUGCGCACAUUAAG -...---...(.(((..(((.--------------.....((((..((...(....).))..))))..((((((((.......)))))))).)))..))).)........ ( -31.10) >consensus GGAC___GGGGG___AUCGAGGAGGAGCUG_______GGAGUUGGGAGU_UGGCAAACUUUUUGGCGAGUGACAUGAAAAGGACAUGUCACACCGUAUGCGCACAUUAAG .......((...............................((((((((..........))))))))..((((((((.......)))))))).))................ (-15.61 = -16.00 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:15 2006