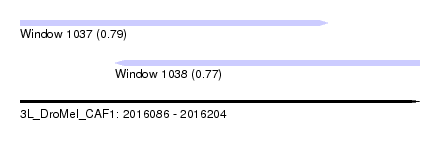
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,016,086 – 2,016,204 |
| Length | 118 |
| Max. P | 0.794722 |

| Location | 2,016,086 – 2,016,177 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 75.27 |
| Mean single sequence MFE | -34.00 |
| Consensus MFE | -18.75 |
| Energy contribution | -21.28 |
| Covariance contribution | 2.53 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
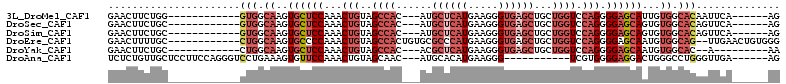
>3L_DroMel_CAF1 2016086 91 + 23771897 GAACUUCUGG------------GUGGCAAGUGCUCCAAACUGUAGCCAC---AUGCUCAUGAAGGGUGAGCUGCUGGUCCAGGGGAGCAUUGUGGCACAAUUCA------AG ..........------------(((.((((((((((...(((..(((((---(.((((((.....)))))))).)))).))).)))))))).)).)))......------.. ( -35.80) >DroSec_CAF1 66621 91 + 1 GAACUUCUGC------------GUGGCAAGUGCUCCAAACUGUAGCCAC---AUGCUCAUGAAGGGUGAGCUGCUGGUCCAGGGGAGCAGUGUGGCACAGUUCA------AG (((((...(.------------((.(((..((((((...(((..(((((---(.((((((.....)))))))).)))).))).)))))).))).)).)))))).------.. ( -37.70) >DroSim_CAF1 81689 91 + 1 GAACUUCUGC------------GUGGCAAGUGCUCCAAACUGUAGCCAC---AUGCUCAUGAAGGGUGAGCUGCUGGUCCAGGGGAGCAGUGUGGCACAGUUCA------AG (((((...(.------------((.(((..((((((...(((..(((((---(.((((((.....)))))))).)))).))).)))))).))).)).)))))).------.. ( -37.70) >DroEre_CAF1 72836 98 + 1 GAACUUUUGC------------CUGGCAAGUGCCCCAAACUGUAGCCACUGUGCGCCCAUGAAGGGUGAGCUGCUGGUCCAGGGGAGCAAUGUGGCAG--UUGAACUGUGGG ..((.(((..------------(((.((..((((((...(((..(((((.((.(((((.....))))).)).).)))).)))))).)))...)).)))--..)))..))... ( -32.30) >DroYak_CAF1 69380 86 + 1 GAACUUCUGC------------CUGGCAAGUGCUCCAAACUGUAGCCAC---ACGCUCAUGAAGGGUGAGCUGCUGGUCCAGGGGAGCAAUGUGGCAC--A---------AA .......(((------------(..(((..((((((...(((..(((((---(.((((((.....)))))))).)))).))).)))))).))))))).--.---------.. ( -33.60) >DroAna_CAF1 64320 92 + 1 UCUCUGUUGCUCCUUCCAGGGUCCUGAAAGUGUUCCAAACUGUAGCAAC---AUGCACAUGAAGGG-----------UCGUGGGGAGGACUGGGCCUGGGUUGA------AG ..........((..((((((.(((....(((.((((........((...---..)).(((((....-----------))))).)))).))))))))))))..))------.. ( -26.90) >consensus GAACUUCUGC____________CUGGCAAGUGCUCCAAACUGUAGCCAC___AUGCUCAUGAAGGGUGAGCUGCUGGUCCAGGGGAGCAAUGUGGCACAGUUCA______AG ......................(((.((..((((((...(((..((((......((((((.....))))))...)))).))).))))))...)).))).............. (-18.75 = -21.28 + 2.53)

| Location | 2,016,114 – 2,016,204 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 78.04 |
| Mean single sequence MFE | -18.37 |
| Consensus MFE | -10.67 |
| Energy contribution | -12.45 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2016114 90 - 23771897 UCC-CCAGUUAUUGUAAUUUACCAUUUCCU------UGAAUUGUGCCACAAUGCUCCCCUGGACCAGCAGCUCACCCUUCAUGAGCAU---GUGGCUACA ...-..........................------.....(((((((((.((((((...)))...)))(((((.......))))).)---))))).))) ( -19.90) >DroSec_CAF1 66649 90 - 1 UCC-CCAGUUAUCGUAAUUUACCAUUUCCU------UGAACUGUGCCACACUGCUCCCCUGGACCAGCAGCUCACCCUUCAUGAGCAU---GUGGCUACA ...-.(((((.....(((.....)))....------..))))).(((((((((.(((...))).)))..(((((.......))))).)---))))).... ( -22.10) >DroSim_CAF1 81717 90 - 1 UCC-CCAGUUAUUGUAAUUUACCAUUUCCU------UGAACUGUGCCACACUGCUCCCCUGGACCAGCAGCUCACCCUUCAUGAGCAU---GUGGCUACA ...-.(((((...(.((........)).).------..))))).(((((((((.(((...))).)))..(((((.......))))).)---))))).... ( -22.70) >DroEre_CAF1 72864 97 - 1 UCU-CCAGUUAUUGUAAUUUACCAUUACCCCACAGUUCAA--CUGCCACAUUGCUCCCCUGGACCAGCAGCUCACCCUUCAUGGGCGCACAGUGGCUACA ...-..(((((((((................)))))..))--))(((((..(((.(((.((((..............)))).))).)))..))))).... ( -20.63) >DroYak_CAF1 69408 85 - 1 CCU-CCAGUUAUUGUAAUUUACCAAUUCUU---------U--GUGCCACAUUGCUCCCCUGGACCAGCAGCUCACCCUUCAUGAGCGU---GUGGCUACA ...-..........................---------(--(((((((((((((((...)))...)))(((((.......)))))))---))))).))) ( -20.50) >DroAna_CAF1 64360 79 - 1 UCAAUUCGUUUUUGUAAUUUAAAAU-UCCU------UCAACCCAGGCCCAGUCCUCCCCACGA-----------CCCUUCAUGUGCAU---GUUGCUACA .....(((((((((.....))))).-....------.......(((......)))....))))-----------.......(((((..---...)).))) ( -4.40) >consensus UCC_CCAGUUAUUGUAAUUUACCAUUUCCU______UGAACUGUGCCACACUGCUCCCCUGGACCAGCAGCUCACCCUUCAUGAGCAU___GUGGCUACA ............................................(((((.(((.((.....)).)))..(((((.......))))).....))))).... (-10.67 = -12.45 + 1.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:20 2006