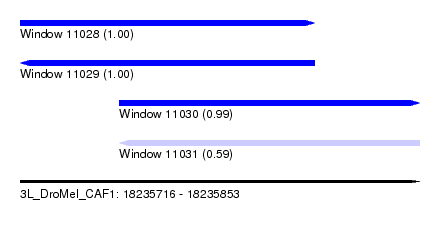
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,235,716 – 18,235,853 |
| Length | 137 |
| Max. P | 0.997276 |

| Location | 18,235,716 – 18,235,817 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 74.61 |
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -9.71 |
| Energy contribution | -11.77 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.36 |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.996198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18235716 101 + 23771897 AUUGCAUUGGUUUUAAAAAUGUUGAACGUGGAGGGGGUUAAUUCCACUCCACAACUGUGCAAAUAUUUGCGAAAACAUUUUCGCAUUUGUAAUGCACAAAA ...(((((.........))))).....((((((.(((.....))).))))))...((((((.(((..(((((((....)))))))..)))..))))))... ( -32.40) >DroSec_CAF1 18333 100 + 1 AUUCCAUGGGUUUUAAAAAUGUUGUGUGUGGAGAGGGUUAAUUCCACUCCACAACUGUGCAAAUAUUUGCGAAAACAUUUUCGCAUUUGUAAUGCACAAA- ..........................(((((((.(((.....))).)))))))..((((((.(((..(((((((....)))))))..)))..))))))..- ( -31.60) >DroSim_CAF1 24356 99 + 1 AUUGCAUUGGUUUUAAAAAUGUUGUGUGUGGAGGGGGUUAAUUCCACUCCACAACUGUGCAAAUAUUUGCGGAAACAUUUUCGCAUUUGUAAUGCACAA-- ((.(((((.........))))).)).(((((((.(((.....))).)))))))..((((((.(((..(((((((....)))))))..)))..)))))).-- ( -32.60) >DroEre_CAF1 18461 94 + 1 AUAUU-CUGGCUUUAAAAAUGUUGAGCGUGGAGAGAGUUAAUAC-----CACAACUGUGCAAAUAUUUGCGAAAACAUUUUCGCAUUUGUAAUGCACAAA- .....-...((((..........))))((((............)-----)))...((((((.(((..(((((((....)))))))..)))..))))))..- ( -23.80) >DroYak_CAF1 25127 95 + 1 AUAGUAUUGGCUUUAAAAAUGUUUUGCGUGGAGGGAGUUAAUCC-----CACAACUGUGCAAAUAUUUGCGAAAACAUUUUCGCAUUUGUAAUGCACAAA- ...((((((.......((((((((.(((..(.((((.....)))-----)....)..)))))))))))((((((....)))))).....)))))).....- ( -25.70) >DroAna_CAF1 23165 80 + 1 ACAAUCCACAUUUGGGAUCAGUUUUUUGUGGCA-UUUUUAAGCC-----ACCAACUGUGCAAAUAUUUCAG-CAUUUUUUUCACAAU-------------- ...((((.(....)))))(((((....(((((.-.......)))-----)).)))))(((..........)-)).............-------------- ( -14.10) >consensus AUAGCAUUGGUUUUAAAAAUGUUGUGCGUGGAGGGGGUUAAUUC_____CACAACUGUGCAAAUAUUUGCGAAAACAUUUUCGCAUUUGUAAUGCACAAA_ .......................................................((((((.(((..(((((((....)))))))..)))..))))))... ( -9.71 = -11.77 + 2.06)


| Location | 18,235,716 – 18,235,817 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 74.61 |
| Mean single sequence MFE | -24.46 |
| Consensus MFE | -8.65 |
| Energy contribution | -10.65 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.35 |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.997276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18235716 101 - 23771897 UUUUGUGCAUUACAAAUGCGAAAAUGUUUUCGCAAAUAUUUGCACAGUUGUGGAGUGGAAUUAACCCCCUCCACGUUCAACAUUUUUAAAACCAAUGCAAU ..(((((((.......(((((((....)))))))......)))))))..((((((.((........))))))))........................... ( -28.12) >DroSec_CAF1 18333 100 - 1 -UUUGUGCAUUACAAAUGCGAAAAUGUUUUCGCAAAUAUUUGCACAGUUGUGGAGUGGAAUUAACCCUCUCCACACACAACAUUUUUAAAACCCAUGGAAU -..((((((.......(((((((....)))))))......))))))(((((((((.((.......)).))))))).))....................... ( -29.82) >DroSim_CAF1 24356 99 - 1 --UUGUGCAUUACAAAUGCGAAAAUGUUUCCGCAAAUAUUUGCACAGUUGUGGAGUGGAAUUAACCCCCUCCACACACAACAUUUUUAAAACCAAUGCAAU --((((((((.....))))(((((((((...((((....))))...(((((((((.((........))))))))).))))))))))).........)))). ( -27.70) >DroEre_CAF1 18461 94 - 1 -UUUGUGCAUUACAAAUGCGAAAAUGUUUUCGCAAAUAUUUGCACAGUUGUG-----GUAUUAACUCUCUCCACGCUCAACAUUUUUAAAGCCAG-AAUAU -..((((((.......(((((((....)))))))......))))))(((.((-----((.((((.....................)))).)))).-))).. ( -20.62) >DroYak_CAF1 25127 95 - 1 -UUUGUGCAUUACAAAUGCGAAAAUGUUUUCGCAAAUAUUUGCACAGUUGUG-----GGAUUAACUCCCUCCACGCAAAACAUUUUUAAAGCCAAUACUAU -..((((((.......(((((((....)))))))......))))))((.(.(-----(((.....)))).).))........................... ( -22.22) >DroAna_CAF1 23165 80 - 1 --------------AUUGUGAAAAAAAUG-CUGAAAUAUUUGCACAGUUGGU-----GGCUUAAAAA-UGCCACAAAAAACUGAUCCCAAAUGUGGAUUGU --------------.............((-(..........)))(((((.((-----(((.......-.)))))....)))))(((((....).))))... ( -18.30) >consensus _UUUGUGCAUUACAAAUGCGAAAAUGUUUUCGCAAAUAUUUGCACAGUUGUG_____GAAUUAACCCCCUCCACACACAACAUUUUUAAAACCAAUACAAU ..(((((((.......(((((((....)))))))......)))))))...................................................... ( -8.65 = -10.65 + 2.00)



| Location | 18,235,750 – 18,235,853 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 90.03 |
| Mean single sequence MFE | -20.58 |
| Consensus MFE | -17.06 |
| Energy contribution | -16.90 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18235750 103 + 23771897 GGGUUAAUUCCACUCCACAACUGUGCAAAUAUUUGCGAAAACAUUUUCGCAUUUGUAAUGCACAAAAAAAAAAUAGUUUCAUUCAAAAAGAGGAUUUU-UUUGU (((.....)))..........((((((.(((..(((((((....)))))))..)))..))))))..((((((((...(((.........))).)))))-))).. ( -21.10) >DroSec_CAF1 18367 97 + 1 GGGUUAAUUCCACUCCACAACUGUGCAAAUAUUUGCGAAAACAUUUUCGCAUUUGUAAUGCACAAA----AAAUAGUUUCAUUCAAAAAGUAGAUUUU-UU--U (((.....)))..........((((((.(((..(((((((....)))))))..)))..))))))..----............................-..--. ( -19.00) >DroSim_CAF1 24390 96 + 1 GGGUUAAUUCCACUCCACAACUGUGCAAAUAUUUGCGGAAACAUUUUCGCAUUUGUAAUGCACAA-----AAAUAGUUUCAUUCAAAAAGUGGAUUUU-UU--U (((.....)))..(((((...((((((.(((..(((((((....)))))))..)))..)))))).-----.....(.......).....)))))....-..--. ( -23.50) >DroEre_CAF1 18494 93 + 1 GAGUUAAUAC-----CACAACUGUGCAAAUAUUUGCGAAAACAUUUUCGCAUUUGUAAUGCACAAA----AAAUAGUUUCAUUCAAAUAGAGGGUUUUUUU--U ........((-----(.....((((((.(((..(((((((....)))))))..)))..))))))..----...........(((.....))))))......--. ( -18.90) >DroYak_CAF1 25161 92 + 1 GAGUUAAUCC-----CACAACUGUGCAAAUAUUUGCGAAAACAUUUUCGCAUUUGUAAUGCACAAA----AAAUAGUUUCAUUCAAAAAGAGGGUUUU-UU--U ........((-----(.....((((((.(((..(((((((....)))))))..)))..))))))..----............((.....)))))....-..--. ( -20.40) >consensus GGGUUAAUUCCACUCCACAACUGUGCAAAUAUUUGCGAAAACAUUUUCGCAUUUGUAAUGCACAAA____AAAUAGUUUCAUUCAAAAAGAGGAUUUU_UU__U .....................((((((.(((..(((((((....)))))))..)))..))))))........................................ (-17.06 = -16.90 + -0.16)

| Location | 18,235,750 – 18,235,853 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 90.03 |
| Mean single sequence MFE | -22.30 |
| Consensus MFE | -14.84 |
| Energy contribution | -17.04 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18235750 103 - 23771897 ACAAA-AAAAUCCUCUUUUUGAAUGAAACUAUUUUUUUUUUGUGCAUUACAAAUGCGAAAAUGUUUUCGCAAAUAUUUGCACAGUUGUGGAGUGGAAUUAACCC .((((-((.......)))))).......((((((..(..(((((((.......(((((((....)))))))......)))))))..)..))))))......... ( -23.72) >DroSec_CAF1 18367 97 - 1 A--AA-AAAAUCUACUUUUUGAAUGAAACUAUUU----UUUGUGCAUUACAAAUGCGAAAAUGUUUUCGCAAAUAUUUGCACAGUUGUGGAGUGGAAUUAACCC .--..-....((((((((((....))))((((..----.(((((((.......(((((((....)))))))......)))))))..))))))))))........ ( -23.32) >DroSim_CAF1 24390 96 - 1 A--AA-AAAAUCCACUUUUUGAAUGAAACUAUUU-----UUGUGCAUUACAAAUGCGAAAAUGUUUCCGCAAAUAUUUGCACAGUUGUGGAGUGGAAUUAACCC .--..-....((((((((((....))))((((..-----(((((((.......((((..........))))......)))))))..))))))))))........ ( -23.22) >DroEre_CAF1 18494 93 - 1 A--AAAAAAACCCUCUAUUUGAAUGAAACUAUUU----UUUGUGCAUUACAAAUGCGAAAAUGUUUUCGCAAAUAUUUGCACAGUUGUG-----GUAUUAACUC .--....................(((.(((((..----.(((((((.......(((((((....)))))))......)))))))..)))-----)).))).... ( -20.92) >DroYak_CAF1 25161 92 - 1 A--AA-AAAACCCUCUUUUUGAAUGAAACUAUUU----UUUGUGCAUUACAAAUGCGAAAAUGUUUUCGCAAAUAUUUGCACAGUUGUG-----GGAUUAACUC .--..-.................(((..((((..----.(((((((.......(((((((....)))))))......)))))))..)))-----)..))).... ( -20.32) >consensus A__AA_AAAAUCCUCUUUUUGAAUGAAACUAUUU____UUUGUGCAUUACAAAUGCGAAAAUGUUUUCGCAAAUAUUUGCACAGUUGUGGAGUGGAAUUAACCC ............................((((((.....(((((((.......(((((((....)))))))......))))))).....))))))......... (-14.84 = -17.04 + 2.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:01 2006