| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,222,829 – 18,222,933 |
| Length | 104 |
| Max. P | 0.733368 |

| Location | 18,222,829 – 18,222,933 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 88.02 |
| Mean single sequence MFE | -20.16 |
| Consensus MFE | -14.66 |
| Energy contribution | -14.94 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733368 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18222829 104 + 23771897 CUUCCCAUUCCCACUUUUAAACCACUGGUCUUUACUUCCUUGAAAUGUGGGCCCACCCACAAAUCUCUCACACUCAGUGUUGUCCGUUG---UGGAUCCAAAAGCAA .............(((((...(((((((.(..((((....(((..((((((....))))))......))).....))))..).)))..)---)))....)))))... ( -20.20) >DroSec_CAF1 5363 100 + 1 CUUCCCAUUCCCACUUUUAAACCACUGGUCUUUACUUCCUUGAAAUGUGGGCCCACCCACAAAUCUCUC----UCAGUGUUGUCAGUUG---UGGAUCCAAAAGCAA ..........((((.........(((((.(..((((.....((..((((((....))))))..))....----..))))..)))))).)---)))............ ( -20.30) >DroSim_CAF1 11835 104 + 1 CUUCCCAUUCCCACUUUUAAACCACUGGUCUUUACUUCCUUGAAAUGUGGGCCCACCCACAAACCUCUCACACUUAGUGUUGUCAGUUG---UGGAUCCAAAAGCAA ..........((((.........(((((.(..((((....(((..((((((....))))))......))).....))))..)))))).)---)))............ ( -19.00) >DroEre_CAF1 5341 104 + 1 CUUCCCAUUCCCACUUUCAAACCACUGGUCUUAACUUCCUUGAAAUGUGGGCCCACCCACAAAUCUUUCAUACUCAGUGUUGUCGGUUG---UGGAACCCAAAACGA ....((((..((.....(((..(((((((...........(((((((((((....))))))....))))).)).))))))))..))..)---)))............ ( -22.20) >DroYak_CAF1 12186 106 + 1 CUUCCCACUCCCUCU-GCAAACCACUGGUCUUUACUUCCUUGAAAUGUGGGCCCACCCACAAAUCCCGCACCCUCAGUGCUGUCAGUUCAGUUAGAUUCAAAAUAAA ...............-..........(((((..(((...((((..((((((....))))))......((((.....))))..))))...))).)))))......... ( -19.10) >consensus CUUCCCAUUCCCACUUUUAAACCACUGGUCUUUACUUCCUUGAAAUGUGGGCCCACCCACAAAUCUCUCACACUCAGUGUUGUCAGUUG___UGGAUCCAAAAGCAA ....(((.....(((..(((..((((((((...........))..((((((....))))))............)))))))))..))).....)))............ (-14.66 = -14.94 + 0.28)

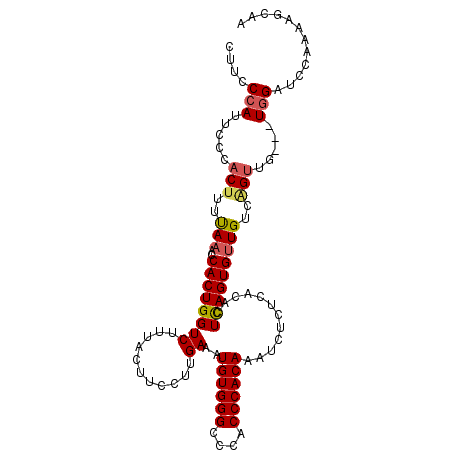

| Location | 18,222,829 – 18,222,933 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 88.02 |
| Mean single sequence MFE | -30.04 |
| Consensus MFE | -21.25 |
| Energy contribution | -21.25 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18222829 104 - 23771897 UUGCUUUUGGAUCCA---CAACGGACAACACUGAGUGUGAGAGAUUUGUGGGUGGGCCCACAUUUCAAGGAAGUAAAGACCAGUGGUUUAAAAGUGGGAAUGGGAAG (..((((((((.(((---(...((....(((.....)))...((..((((((....))))))..)).............)).))))))))))))..).......... ( -31.00) >DroSec_CAF1 5363 100 - 1 UUGCUUUUGGAUCCA---CAACUGACAACACUGA----GAGAGAUUUGUGGGUGGGCCCACAUUUCAAGGAAGUAAAGACCAGUGGUUUAAAAGUGGGAAUGGGAAG (..((((((((.(((---(...............----....((..((((((....))))))..))..((.........)).))))))))))))..).......... ( -28.30) >DroSim_CAF1 11835 104 - 1 UUGCUUUUGGAUCCA---CAACUGACAACACUAAGUGUGAGAGGUUUGUGGGUGGGCCCACAUUUCAAGGAAGUAAAGACCAGUGGUUUAAAAGUGGGAAUGGGAAG (..((((((((.(((---(..((.(((........))).)).((((((((((....)))))(((((...)))))..))))).))))))))))))..).......... ( -32.70) >DroEre_CAF1 5341 104 - 1 UCGUUUUGGGUUCCA---CAACCGACAACACUGAGUAUGAAAGAUUUGUGGGUGGGCCCACAUUUCAAGGAAGUUAAGACCAGUGGUUUGAAAGUGGGAAUGGGAAG ...((((.(.(((((---(...(((...(((((.........((..((((((....))))))..))......((....)))))))..)))...)))))).).)))). ( -27.80) >DroYak_CAF1 12186 106 - 1 UUUAUUUUGAAUCUAACUGAACUGACAGCACUGAGGGUGCGGGAUUUGUGGGUGGGCCCACAUUUCAAGGAAGUAAAGACCAGUGGUUUGC-AGAGGGAGUGGGAAG ....((((.(.(((..(((.(((..(.((((.....))))..((..((((((....))))))..))..)..)))..(((((...))))).)-))..))).).)))). ( -30.40) >consensus UUGCUUUUGGAUCCA___CAACUGACAACACUGAGUGUGAGAGAUUUGUGGGUGGGCCCACAUUUCAAGGAAGUAAAGACCAGUGGUUUAAAAGUGGGAAUGGGAAG ...(((((.(.(((.........(((..(((((.........((..((((((....))))))..))......((....))))))))))........))).).))))) (-21.25 = -21.25 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:53 2006