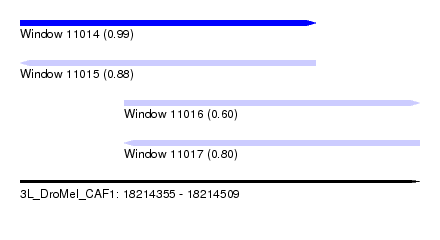
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,214,355 – 18,214,509 |
| Length | 154 |
| Max. P | 0.985043 |

| Location | 18,214,355 – 18,214,469 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 93.27 |
| Mean single sequence MFE | -38.77 |
| Consensus MFE | -35.91 |
| Energy contribution | -37.47 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.985043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18214355 114 + 23771897 UAUUUUGUUCUCUGUAUUCAAAAUAGGCAGCAAGAUAAAAGCCGUAACAGGCCUUGUGGCGAGCCUUUAACAUUUGUUAAAGCCACGAGUGCCAUUCGGCCUCUUUGGGCUCUG (((((((((.(((((.......))))).)))))))))..((((..((.(((((..((((((..((((((((....)))))))....)..))))))..))))).))..))))... ( -39.50) >DroSec_CAF1 115826 114 + 1 UAUUUUCCUCUCUGUAGUCAAAAUAGGCAAAAAGAUAAAAGCCCAUGCAGGCCUUGUGGCGAGCCUUUAACAUUUGUUAAAGCCACGAGUGCCAUUCGGCCUCUUUGGGCUCUG ..........(((...(((......)))....)))....((((((...(((((..((((((..((((((((....)))))))....)..))))))..)))))...))))))... ( -37.80) >DroEre_CAF1 118007 114 + 1 CAUUUUGUUCUCUGUAGUCAAAAUAGGCAAUAAGAAAAAAGCCGAAGCAGGCCUUGUGGCGAGCCUUUAACAUUUGUUAAAGCCACGAGUGCCAUUCGGCCUCUUUGGGCUCUG ..(((((((.(((((.......))))).)))))))....((((.(((.(((((..((((((..((((((((....)))))))....)..))))))..))))).))).))))... ( -39.00) >consensus UAUUUUGUUCUCUGUAGUCAAAAUAGGCAAAAAGAUAAAAGCCGAAGCAGGCCUUGUGGCGAGCCUUUAACAUUUGUUAAAGCCACGAGUGCCAUUCGGCCUCUUUGGGCUCUG (((((((((.(((((.......))))).)))))))))..((((.(((.(((((..((((((..((((((((....)))))))....)..))))))..))))).))).))))... (-35.91 = -37.47 + 1.56)



| Location | 18,214,355 – 18,214,469 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 93.27 |
| Mean single sequence MFE | -35.50 |
| Consensus MFE | -30.70 |
| Energy contribution | -30.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18214355 114 - 23771897 CAGAGCCCAAAGAGGCCGAAUGGCACUCGUGGCUUUAACAAAUGUUAAAGGCUCGCCACAAGGCCUGUUACGGCUUUUAUCUUGCUGCCUAUUUUGAAUACAGAGAACAAAAUA .((((((..((.(((((...((((....(.(.(((((((....))))))).).)))))...))))).))..))))))............(((((((...........))))))) ( -33.00) >DroSec_CAF1 115826 114 - 1 CAGAGCCCAAAGAGGCCGAAUGGCACUCGUGGCUUUAACAAAUGUUAAAGGCUCGCCACAAGGCCUGCAUGGGCUUUUAUCUUUUUGCCUAUUUUGACUACAGAGAGGAAAAUA .((((((((...(((((...((((....(.(.(((((((....))))))).).)))))...)))))...)))))))).....((((.(((.(((((....)))))))))))).. ( -39.80) >DroEre_CAF1 118007 114 - 1 CAGAGCCCAAAGAGGCCGAAUGGCACUCGUGGCUUUAACAAAUGUUAAAGGCUCGCCACAAGGCCUGCUUCGGCUUUUUUCUUAUUGCCUAUUUUGACUACAGAGAACAAAAUG .((.((..((((((((((((.(((.((.(((((((((((....)))))).....))))).)))))...))))))))))))......))))((((((.((....))..)))))). ( -33.70) >consensus CAGAGCCCAAAGAGGCCGAAUGGCACUCGUGGCUUUAACAAAUGUUAAAGGCUCGCCACAAGGCCUGCUUCGGCUUUUAUCUUAUUGCCUAUUUUGACUACAGAGAACAAAAUA .((((((.....(((((...((((....(.(.(((((((....))))))).).)))))...))))).....))))))..............(((((....)))))......... (-30.70 = -30.70 + -0.00)


| Location | 18,214,395 – 18,214,509 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.76 |
| Mean single sequence MFE | -36.78 |
| Consensus MFE | -24.62 |
| Energy contribution | -24.10 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18214395 114 + 23771897 GCCGUAACAGGCCUUGUGGCGAGCCU-UUAACAUUUGUUAAAGCCACGAGUGCCAUUCGGCCUCUUUGGGCU-----CUGUGCAUACUGCAGGUAAUUUUCGCAACGACGCAAUUGAGUU ((((.....(((((((((((.....(-(((((....)))))))))))))).)))...)))).......((((-----(..(((....(((..(.....)..))).....)))...))))) ( -36.70) >DroPse_CAF1 146282 106 + 1 ----------GCCUUGUGGCGAGCCUUUUAACAUUUGU----GCCACAAGUGGCUCUCGGCCUCUUUGGGCUCUGCUCUGUGCAUAUUGCAGUUAAUUUUCGCAACGACGCAAUAGAGUU ----------(((....)))((((((..........(.----((((....)))).)...........)))))).(((((((((...((((...........))))....)).))))))). ( -32.40) >DroSec_CAF1 115866 114 + 1 GCCCAUGCAGGCCUUGUGGCGAGCCU-UUAACAUUUGUUAAAGCCACGAGUGCCAUUCGGCCUCUUUGGGCU-----CUGUGCAUACUGCAGGUAAUUUUCGCAACGACGCAAUUGAGUU (((((...(((((..((((((..(((-(((((....)))))))....)..))))))..)))))...)))))(-----(..(((....(((..(.....)..))).....)))...))... ( -39.40) >DroEre_CAF1 118047 114 + 1 GCCGAAGCAGGCCUUGUGGCGAGCCU-UUAACAUUUGUUAAAGCCACGAGUGCCAUUCGGCCUCUUUGGGCU-----CUGUGCAUGCUGCAGGUAAUUUUCGCAACUACGCAAUUGAGUU .((((((.(((((..((((((..(((-(((((....)))))))....)..))))))..)))))))))))(((-----(..(((..(.(((..(.....)..))).)...)))...)))). ( -43.00) >DroPer_CAF1 135413 106 + 1 ----------GCCUUGUGGCGAGCCUUUUAACAUUUGU----GCCACAAGUGGCUCUCGGCCUCUUUGGGCUCUGCUCUGUGCAUAUUGCAGUUAAUUUUCGCAACGACGCAAUAGAGUU ----------(((....)))((((((..........(.----((((....)))).)...........)))))).(((((((((...((((...........))))....)).))))))). ( -32.40) >consensus GCC____CAGGCCUUGUGGCGAGCCU_UUAACAUUUGUUAAAGCCACGAGUGCCAUUCGGCCUCUUUGGGCU_____CUGUGCAUACUGCAGGUAAUUUUCGCAACGACGCAAUUGAGUU ..........((.((((.(((((........(((((((.......)))))))((....(((((....)))))........(((.....))))).....))))).)))).))......... (-24.62 = -24.10 + -0.52)


| Location | 18,214,395 – 18,214,509 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.76 |
| Mean single sequence MFE | -35.06 |
| Consensus MFE | -24.23 |
| Energy contribution | -23.99 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18214395 114 - 23771897 AACUCAAUUGCGUCGUUGCGAAAAUUACCUGCAGUAUGCACAG-----AGCCCAAAGAGGCCGAAUGGCACUCGUGGCUUUAACAAAUGUUAA-AGGCUCGCCACAAGGCCUGUUACGGC ........(((((.((((((.........)))))))))))...-----.(((..((.(((((...((((....(.(.(((((((....)))))-)).).)))))...))))).))..))) ( -34.00) >DroPse_CAF1 146282 106 - 1 AACUCUAUUGCGUCGUUGCGAAAAUUAACUGCAAUAUGCACAGAGCAGAGCCCAAAGAGGCCGAGAGCCACUUGUGGC----ACAAAUGUUAAAAGGCUCGCCACAAGGC---------- ..((((..(((((.((((((.........))))))))))).))))..(((((..(((.(((.....))).))).((((----(....)))))...)))))(((....)))---------- ( -31.30) >DroSec_CAF1 115866 114 - 1 AACUCAAUUGCGUCGUUGCGAAAAUUACCUGCAGUAUGCACAG-----AGCCCAAAGAGGCCGAAUGGCACUCGUGGCUUUAACAAAUGUUAA-AGGCUCGCCACAAGGCCUGCAUGGGC .......(((((....)))))......(((((((........(-----((((((..((((((....))).))).)))(((((((....)))))-))))))(((....)))))))).)).. ( -38.50) >DroEre_CAF1 118047 114 - 1 AACUCAAUUGCGUAGUUGCGAAAAUUACCUGCAGCAUGCACAG-----AGCCCAAAGAGGCCGAAUGGCACUCGUGGCUUUAACAAAUGUUAA-AGGCUCGCCACAAGGCCUGCUUCGGC ..(((...(((((.((((((.........)))))))))))..)-----)).((.((((((((...((((....(.(.(((((((....)))))-)).).)))))...))))).))).)). ( -40.20) >DroPer_CAF1 135413 106 - 1 AACUCUAUUGCGUCGUUGCGAAAAUUAACUGCAAUAUGCACAGAGCAGAGCCCAAAGAGGCCGAGAGCCACUUGUGGC----ACAAAUGUUAAAAGGCUCGCCACAAGGC---------- ..((((..(((((.((((((.........))))))))))).))))..(((((..(((.(((.....))).))).((((----(....)))))...)))))(((....)))---------- ( -31.30) >consensus AACUCAAUUGCGUCGUUGCGAAAAUUACCUGCAGUAUGCACAG_____AGCCCAAAGAGGCCGAAUGGCACUCGUGGCUUUAACAAAUGUUAA_AGGCUCGCCACAAGGCCUG____GGC ........(((((.((((((.........))))))))))).........(((....((((((....))).)))(((((......................)))))..))).......... (-24.23 = -23.99 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:47 2006