
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,190,834 – 18,190,924 |
| Length | 90 |
| Max. P | 0.759160 |

| Location | 18,190,834 – 18,190,924 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 80.64 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -22.58 |
| Energy contribution | -22.67 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
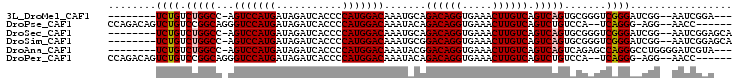
>3L_DroMel_CAF1 18190834 90 - 23771897 --------UCUGUCUGGCC-AGUCCAUGAUAGAUCACCCCAUGGACAAAUGCAGACAGGUGAAACUUGUCAGUCAGUGCGGGUCGGGAUCGG--AAUCGGA--- --------(((((((((((-.(((((((...........)))))))...((((((((((.....))))))......)))))))))))..)))--)......--- ( -30.60) >DroPse_CAF1 123898 93 - 1 CCAGACAGUCUGUCCGGCAGGGUCCAUGAUAGAUCACCCCAUGGACAAAUACAGACAGGUGAAACUUGUCAGUCUGUCCA--UCAGGG-AGG--AACC------ ((.(((.....))).))..((.(((.(((....)))(((.(((((((...((.((((((.....)))))).)).))))))--)..)))-.))--).))------ ( -30.10) >DroSec_CAF1 92829 93 - 1 --------UCUGUCUGGCC-AGUCCAUGAUAGAUCACCCCAUGGACAAAUGCAGACAGGUGAAACUUGUCAGUCAGUGCGGGUCGGGAUCGG--AAUCGGAGCA --------(((((((((((-.(((((((...........)))))))...((((((((((.....))))))......)))))))))))..)))--)......... ( -30.60) >DroSim_CAF1 93516 93 - 1 --------UCUGUCUGGCC-AGUCCAUGAUAGAUCACCCCAUGGACAAAUGCGGACAGGUGAAACUUGUCAGUCAGUGCGGGUCGGGAUCGG--AAUCGGAGCA --------(((((((((((-.(((((((...........)))))))....((.((((((.....)))))).)).......)))))))..)))--)......... ( -29.90) >DroAna_CAF1 101629 92 - 1 --------UCUGUCUGGCC-AGUCCAUGAUAGAUCACCCCAUGGACAAAUACGGACAGGUGAAACUUGUCAGUCAGUCAGAGCCAGGGCCUGGGGAUCGUA--- --------((((.(((((.-.(((((((...........))))))).......((((((.....)))))).))))).)))).((((...))))........--- ( -32.90) >DroPer_CAF1 113010 93 - 1 CCAGACAGUCUGUCCGGCAGGGUCCAUGAUAGAUCACCCCAUGGACAAAUACAGACAGGUGAAACUUGUCAGUCUGUCCA--UCAGGG-AGG--AACC------ ((.(((.....))).))..((.(((.(((....)))(((.(((((((...((.((((((.....)))))).)).))))))--)..)))-.))--).))------ ( -30.10) >consensus ________UCUGUCUGGCC_AGUCCAUGAUAGAUCACCCCAUGGACAAAUACAGACAGGUGAAACUUGUCAGUCAGUCCGGGUCAGGAUCGG__AAUCGGA___ ........((((.(((((...(((((((...........))))))).......((((((.....)))))).))))).......))))................. (-22.58 = -22.67 + 0.08)
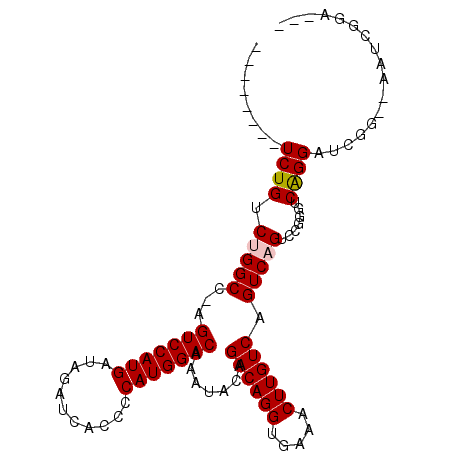
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:38 2006