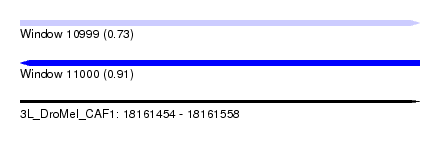
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,161,454 – 18,161,558 |
| Length | 104 |
| Max. P | 0.909130 |

| Location | 18,161,454 – 18,161,558 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.30 |
| Mean single sequence MFE | -36.90 |
| Consensus MFE | -22.36 |
| Energy contribution | -21.45 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
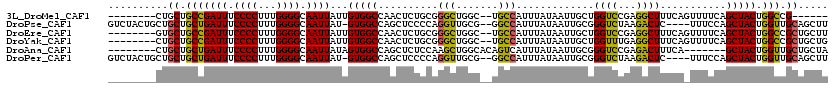
>3L_DroMel_CAF1 18161454 104 + 23771897 --------CUGCUGCCGAUUUCCCCUUUGGGGCAAUUAUUGUGGCCAACUCUGCGGGCUGGC--UGCCAUUUAUAAUUGCUGGUCCGAGGCUUUCAGUUUUCAGCUACUGGCCG------ --------..((..(.((((.((((...)))).))))...)..))........(((((..((--..............))..))))).((((...(((.....)))...)))).------ ( -32.94) >DroPse_CAF1 88270 113 + 1 GUCUACUGCUGCUGCUGAUUUCCCCUUUGGGGCAAUUAU-GUGGCCAGCUCCCCAGGUUGCG--GGCCAUUUAUAAUUGCGGGUCUAAGACUC----UUUCCAGCUACUGGUUGCAGCUU .......(((((.((((....(((....)))((((((((-((((((.((..(....)..)).--))))))..))))))))(((((...)))))----....))))........))))).. ( -41.80) >DroEre_CAF1 64742 110 + 1 --------GUGCUGCCGAUUUCCCCUUUGGGGCAAUUAUUGUGGCCAACUCUGCGGGCUGGC--UGCCAUUUAUAAUUGCUGGUCCGAGGCUUUCAGUUUUCAGCUACUGGCCGCUGCUU --------..((....((((.((((...)))).))))...(((((((......(((((..((--..............))..))))).((((..........))))..))))))).)).. ( -36.54) >DroYak_CAF1 65758 110 + 1 --------CUGCUGCCGAUUUCCCCUUUGGGGCAAUUAUUGUGGCCAACUCUGCGGGCUGGC--UGCCAUUUAUAAUUGCUGGUUUGAGGCUUUCAGUUUUCAGCUACUGGCCGCUGCUG --------..((.((((....(((....)))((((((((.(..((((.((.....)).))))--..).....))))))))(((((.(((((.....))))).))))).)))).))..... ( -34.80) >DroAna_CAF1 70307 105 + 1 --------CUGCUGCUGAUUUCCCCUUUGGGGCAAUUAUAGUGGCCAGCUCUCCAAGCUGGCACAGUCAUUUAUAAUUGCGGGUCCGAGACUUUCA-------GCUACUGGUUGCUGCUA --------.....(((((...(((....)))(((((((((((((((((((.....))))))).)).....))))))))))(((((...))))))))-------))...((((....)))) ( -33.50) >DroPer_CAF1 76827 113 + 1 GUCUACUGCUGCUGCUGAUUUCCCCUUUGGGGCAAUUAU-GUGGCCAGCUCCCCAGGUUGCG--GGCCAUUUAUAAUUGCGGGUCUAAGACUC----UUUCCAGCUACUGGUUGCAGCUU .......(((((.((((....(((....)))((((((((-((((((.((..(....)..)).--))))))..))))))))(((((...)))))----....))))........))))).. ( -41.80) >consensus ________CUGCUGCCGAUUUCCCCUUUGGGGCAAUUAUUGUGGCCAACUCUCCAGGCUGGC__UGCCAUUUAUAAUUGCGGGUCCGAGACUUUCA_UUUUCAGCUACUGGCCGCUGCUU ..........((.(((((((.((((...)))).))))...(((((..........(((.......))).............((((...))))...........))))).))).))..... (-22.36 = -21.45 + -0.91)

| Location | 18,161,454 – 18,161,558 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.30 |
| Mean single sequence MFE | -34.13 |
| Consensus MFE | -22.43 |
| Energy contribution | -23.02 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.909130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18161454 104 - 23771897 ------CGGCCAGUAGCUGAAAACUGAAAGCCUCGGACCAGCAAUUAUAAAUGGCA--GCCAGCCCGCAGAGUUGGCCACAAUAAUUGCCCCAAAGGGGAAAUCGGCAGCAG-------- ------.........((((....((((...((((......((((((((...((...--((((((.......))))))..))))))))))......))))...))))))))..-------- ( -30.00) >DroPse_CAF1 88270 113 - 1 AAGCUGCAACCAGUAGCUGGAAA----GAGUCUUAGACCCGCAAUUAUAAAUGGCC--CGCAACCUGGGGAGCUGGCCAC-AUAAUUGCCCCAAAGGGGAAAUCAGCAGCAGCAGUAGAC ..(((((..((((...))))...----((...........((((((((...(((((--.((..(....)..)).))))).-))))))))(((....)))...)).))))).......... ( -38.20) >DroEre_CAF1 64742 110 - 1 AAGCAGCGGCCAGUAGCUGAAAACUGAAAGCCUCGGACCAGCAAUUAUAAAUGGCA--GCCAGCCCGCAGAGUUGGCCACAAUAAUUGCCCCAAAGGGGAAAUCGGCAGCAC-------- ..((.(((((((((........))))...))).(((....((((((((...((...--((((((.......))))))..))))))))))(((....)))...))))).))..-------- ( -31.60) >DroYak_CAF1 65758 110 - 1 CAGCAGCGGCCAGUAGCUGAAAACUGAAAGCCUCAAACCAGCAAUUAUAAAUGGCA--GCCAGCCCGCAGAGUUGGCCACAAUAAUUGCCCCAAAGGGGAAAUCGGCAGCAG-------- ((((.((.....)).))))....(((...(((........((((((((...((...--((((((.......))))))..))))))))))(((....))).....)))..)))-------- ( -33.10) >DroAna_CAF1 70307 105 - 1 UAGCAGCAACCAGUAGC-------UGAAAGUCUCGGACCCGCAAUUAUAAAUGACUGUGCCAGCUUGGAGAGCUGGCCACUAUAAUUGCCCCAAAGGGGAAAUCAGCAGCAG-------- ..((.((.....)).((-------(((...((((......(((((((((......((.((((((((...)))))))))).)))))))))......))))...))))).))..-------- ( -33.70) >DroPer_CAF1 76827 113 - 1 AAGCUGCAACCAGUAGCUGGAAA----GAGUCUUAGACCCGCAAUUAUAAAUGGCC--CGCAACCUGGGGAGCUGGCCAC-AUAAUUGCCCCAAAGGGGAAAUCAGCAGCAGCAGUAGAC ..(((((..((((...))))...----((...........((((((((...(((((--.((..(....)..)).))))).-))))))))(((....)))...)).))))).......... ( -38.20) >consensus AAGCAGCAACCAGUAGCUGAAAA_UGAAAGCCUCAGACCAGCAAUUAUAAAUGGCA__GCCAGCCCGCAGAGCUGGCCACAAUAAUUGCCCCAAAGGGGAAAUCAGCAGCAG________ ............((.(((((....................((((((((...((((.....((((.......))))))))..))))))))(((....)))...))))).)).......... (-22.43 = -23.02 + 0.58)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:31 2006