| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
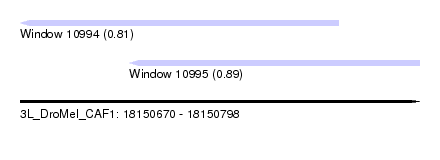
| Location | 18,150,670 – 18,150,798 |
| Length | 128 |
| Max. P | 0.892237 |

| Location | 18,150,670 – 18,150,772 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 76.64 |
| Mean single sequence MFE | -17.24 |
| Consensus MFE | -7.59 |
| Energy contribution | -7.45 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
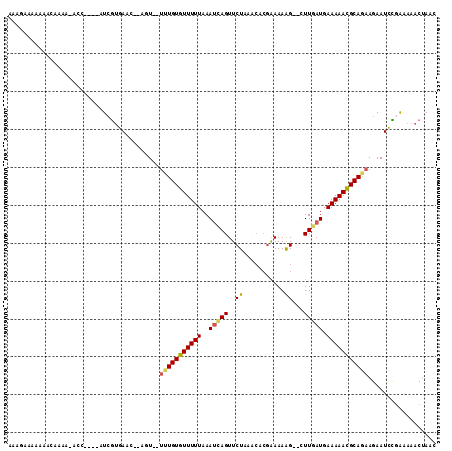
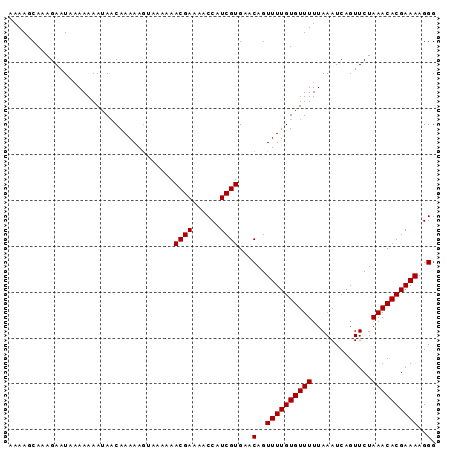
>3L_DroMel_CAF1 18150670 102 - 23771897 AAAAUAAAAAACGAAA-UCC----AUCGUGAAC--AGU--UUUGUGUUUUUAAAUCAGUUCUAAACACGAAAAGG--GUUGAUGAAAAACGCAGAAGAAUCCGAAAAACUAAC ................-...----.(((.((..--..(--(((((((((((..(((((..((..........)).--.))))).))))))))))))...)))))......... ( -20.00) >DroSec_CAF1 53241 102 - 1 AAAGUAAAAAACGAAA-ACC----AUCGUGAAC--AGU--UUUGUGUUUUUAAAUCAGUUCUAAACACGAAAAGG--GUUGAUGAAAAACGCAGAAGAAUCCGAAAAACUAAC ..(((.....((((..-...----.))))....--..(--(((((((((((..(((((..((..........)).--.))))).))))))))))))...........)))... ( -20.40) >DroSim_CAF1 50696 102 - 1 AAAGAAAAAAACGAAA-ACC----AUCGUGAAC--AGU--UUUGUGUUUUUAAAUCAGUUCUAAACACGAAAAGG--GUUGAUGAAAAACGCAGAAGAAUCCGAAAAACUAAC ................-...----.(((.((..--..(--(((((((((((..(((((..((..........)).--.))))).))))))))))))...)))))......... ( -20.00) >DroYak_CAF1 55251 106 - 1 GUAAAAAAAAACAAAA-AUC----AUCGUGACAAAAAUUGUUUGUGUUUUUAAAUCAGUUCUAAACACGAAAAAG--CUUGAUGAAAAACGCAGAAGAAUCCGAAAAACUAAC ................-...----.(((.((.........(((((((((((..(((((((.............))--).)))).)))))))))))....)))))......... ( -12.64) >DroAna_CAF1 62835 102 - 1 GUAGAAGAAUUAAAAA-AAUAUAUUUGGUGAUU--UUU--GGUGUGUUUUUAAAGAAGUUCUAACC---GAAAAG--CUUGAUGAAAAAUGCAAAAGAAUCUGGAAAACUAA- .(((....((((((..-......))))))((((--(((--..(((((((((.(((...(((.....---)))...--)))....))))))))).))))))).......))).- ( -15.70) >DroPer_CAF1 68494 96 - 1 ----AAUAAAACAAACCCCC----AUCCCGAUA--UUU--UUUGUGUUUUUAAAUAAGUUCU-----CGAAAAAGUUCUUUAUGAAAAAUGCAAAAGAAUUGAAAAAACAAAA ----................----....((((.--(((--(((((((((((....(((..((-----......))..)))....))))))))))))))))))........... ( -14.70) >consensus AAAGAAAAAAACAAAA_ACC____AUCGUGAAC__AGU__UUUGUGUUUUUAAAUCAGUUCUAAACACGAAAAAG__CUUGAUGAAAAACGCAGAAGAAUCCGAAAAACUAAC ........................................(((((((((((..(((((..((...........))...))))).))))))))))).................. ( -7.59 = -7.45 + -0.14)


| Location | 18,150,705 – 18,150,798 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 95.70 |
| Mean single sequence MFE | -11.52 |
| Consensus MFE | -11.22 |
| Energy contribution | -11.22 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
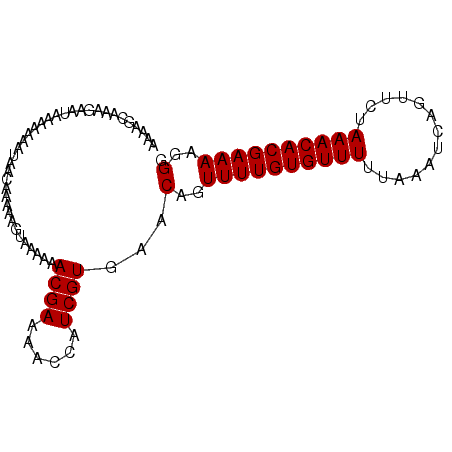
>3L_DroMel_CAF1 18150705 93 - 23771897 AAAAGCAAAGAAUAAAAAAAUAACAAAAAAUAAAAAACGAAAUCCAUCGUGAACAGUUUUGUGUUUUUAAAUCAGUUCUAAACACGAAAAGGG ....................................((((......))))...(..((((((((((.............))))))))))..). ( -11.22) >DroSec_CAF1 53276 93 - 1 AAAAGCAAAGAAUAAAAAAAAACCAAAAAGUAAAAAACGAAAACCAUCGUGAACAGUUUUGUGUUUUUAAAUCAGUUCUAAACACGAAAAGGG ......................((............((((......))))......((((((((((.............)))))))))).)). ( -12.12) >DroSim_CAF1 50731 93 - 1 AAAAGCAAAGAAUACAAAAAUAACAAAAAGAAAAAAACGAAAACCAUCGUGAACAGUUUUGUGUUUUUAAAUCAGUUCUAAACACGAAAAGGG ....................................((((......))))...(..((((((((((.............))))))))))..). ( -11.22) >consensus AAAAGCAAAGAAUAAAAAAAUAACAAAAAGUAAAAAACGAAAACCAUCGUGAACAGUUUUGUGUUUUUAAAUCAGUUCUAAACACGAAAAGGG ....................................((((......))))...(..((((((((((.............))))))))))..). (-11.22 = -11.22 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:25 2006