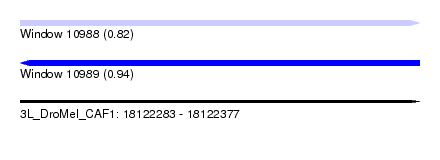
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,122,283 – 18,122,377 |
| Length | 94 |
| Max. P | 0.940286 |

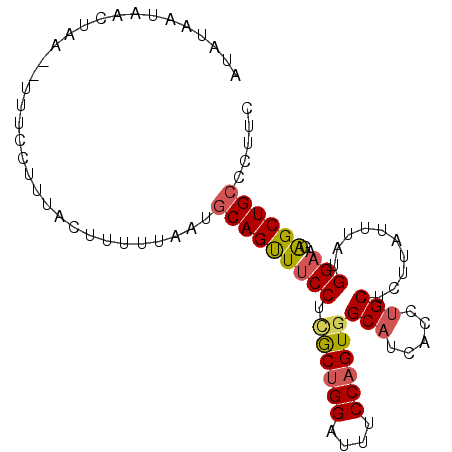
| Location | 18,122,283 – 18,122,377 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 82.25 |
| Mean single sequence MFE | -23.35 |
| Consensus MFE | -18.35 |
| Energy contribution | -18.13 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.819840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18122283 94 + 23771897 GAAGUGCAGCUCUUCCAUAAAUAAGAGCAGGUGAUGCUACUGGAAAUCCAGCGAGGAAACUGCAUUAAAAGGUAAAUAGAA--UAAGUUAUUAUAU ..(((((((.(((((..........((((.....)))).((((....)))).)))))..)))))))...............--............. ( -21.70) >DroPse_CAF1 34678 96 + 1 GCAGGGCAGCCCUGCCGUAGAUCAGGGCGGGCGAGGCCACGGGAAAGCCAGUGAGGAAGCUGUUUGGAAAAGGCAAGGAAAAUUUAGUAAAAGUAU ((..(((.((((.(((.(.....).)))))))...))).........((((..((....))..)))).....))...................... ( -25.80) >DroSec_CAF1 25324 94 + 1 GAAGGGCAGCUCUUCCAUAAAUAAGAGCAGGUGAUGCCACUGGAAAUCCAGCAAGGAAACUGCAUUAAGAAGUAAAUGAAA--UUAGUUAUUAUAU ....(((.((((((........))))))...((((((..((((....))))..((....))))))))..............--...)))....... ( -21.00) >DroSim_CAF1 22880 94 + 1 GAAGGGCAGCUCUUCCAUAAAUAAGAGCAGGUGAUGCCACUGGAAAUCCAGCGAGGAAACUGCAUUAAAAGGUAAAAGAAA--UUUGUCAUUAAAU ....((((((((((........))))))...(((((((.((((....)))).)((....))))))))..............--..))))....... ( -24.80) >DroEre_CAF1 26121 92 + 1 GGAGGGCAGCUCUUCCAUAAAUGAGAGCAGGUGAUGCCACUGGAAAUCCAGCGAGGAAGCUGCAUU--AAAGUAAAGGAAG--CUCGUUAUUAUAU ...((((.((((((........))))))...(((((((.((((....)))).)((....)))))))--)...........)--))).......... ( -25.20) >DroYak_CAF1 26055 94 + 1 GAAGGGCAGCUCUUCCGUAAAUGAGAGCAGGUGAUGCCACUGGAAAGCCAGCGAGGAAACUGCAAUUAAAAGUUAAGGAAA--UUAGUUAUUAUAU ....(((.((((((........)))))).((((....)))).....))).((.((....))))..................--............. ( -21.60) >consensus GAAGGGCAGCUCUUCCAUAAAUAAGAGCAGGUGAUGCCACUGGAAAUCCAGCGAGGAAACUGCAUUAAAAAGUAAAGGAAA__UUAGUUAUUAUAU ....(((.((((((........))))))...((((((..((((....))))..((....))))))))...................)))....... (-18.35 = -18.13 + -0.22)

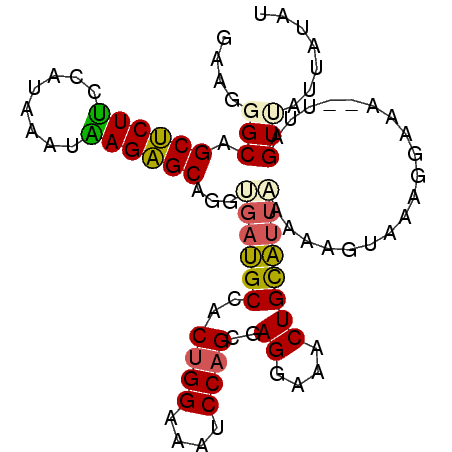
| Location | 18,122,283 – 18,122,377 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 82.25 |
| Mean single sequence MFE | -20.43 |
| Consensus MFE | -17.76 |
| Energy contribution | -17.98 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.940286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18122283 94 - 23771897 AUAUAAUAACUUA--UUCUAUUUACCUUUUAAUGCAGUUUCCUCGCUGGAUUUCCAGUAGCAUCACCUGCUCUUAUUUAUGGAAGAGCUGCACUUC .............--.................((((((((((..(((((....)))))((((.....)))).........))))..)))))).... ( -21.00) >DroPse_CAF1 34678 96 - 1 AUACUUUUACUAAAUUUUCCUUGCCUUUUCCAAACAGCUUCCUCACUGGCUUUCCCGUGGCCUCGCCCGCCCUGAUCUACGGCAGGGCUGCCCUGC ..................(((((((..........((((........))))....((.(((...)))))...........)))))))......... ( -18.50) >DroSec_CAF1 25324 94 - 1 AUAUAAUAACUAA--UUUCAUUUACUUCUUAAUGCAGUUUCCUUGCUGGAUUUCCAGUGGCAUCACCUGCUCUUAUUUAUGGAAGAGCUGCCCUUC .............--..................(((((((((..(((((....)))))((((.....)))).........))))..)))))..... ( -19.30) >DroSim_CAF1 22880 94 - 1 AUUUAAUGACAAA--UUUCUUUUACCUUUUAAUGCAGUUUCCUCGCUGGAUUUCCAGUGGCAUCACCUGCUCUUAUUUAUGGAAGAGCUGCCCUUC .............--..................(((((((((.((((((....))))))(((.....)))..........))))..)))))..... ( -21.00) >DroEre_CAF1 26121 92 - 1 AUAUAAUAACGAG--CUUCCUUUACUUU--AAUGCAGCUUCCUCGCUGGAUUUCCAGUGGCAUCACCUGCUCUCAUUUAUGGAAGAGCUGCCCUCC .............--.............--...(((((((((.((((((....))))))(((.....)))..........)))..))))))..... ( -23.10) >DroYak_CAF1 26055 94 - 1 AUAUAAUAACUAA--UUUCCUUAACUUUUAAUUGCAGUUUCCUCGCUGGCUUUCCAGUGGCAUCACCUGCUCUCAUUUACGGAAGAGCUGCCCUUC .............--..................(((((((((.((((((....))))))(((.....)))..........))))..)))))..... ( -19.70) >consensus AUAUAAUAACUAA__UUUCCUUUACUUUUUAAUGCAGUUUCCUCGCUGGAUUUCCAGUGGCAUCACCUGCUCUUAUUUAUGGAAGAGCUGCCCUUC .................................(((((((((.((((((....))))))(((.....)))..........)))..))))))..... (-17.76 = -17.98 + 0.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:19 2006