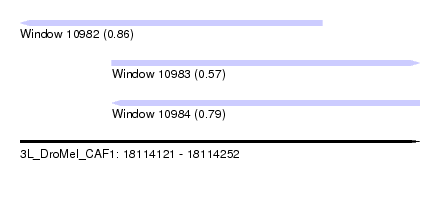
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,114,121 – 18,114,252 |
| Length | 131 |
| Max. P | 0.859249 |

| Location | 18,114,121 – 18,114,220 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 76.66 |
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -17.60 |
| Energy contribution | -17.80 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18114121 99 - 23771897 CAGGCGACUUGGUGCGCUUAAAGUCUUCGACUGACUGAAGCGACCUGCUGCCGCUUUAAGCCGCAAAAGCACCGCAUAAAAAACAAAAA-ACAAG-CAUAC ...(((....(((.(((((..((((.......)))).))))))))((((((.((.....)).))...)))).)))..............-.....-..... ( -24.50) >DroVir_CAF1 18207 92 - 1 UAGGCAACUUGGUGCGUUUAAAGUCUUCGAAUGACUGACGCGACUUGGGGCCGCCUCAAGCCGCCAAAGCGCGGCAAACAA-------A-AAACU-AAACC ..(((..(..(.((((((...((((.......)))))))))).)..)..))).......(((((......)))))......-------.-.....-..... ( -28.70) >DroGri_CAF1 20980 93 - 1 GCAACAACUUGGUGCGCUUCAGGUCUUUGAGUGACUGAAGCGACUUGGGUCCGCUUCGAGCCGCCAAAGCGCAGCAGACACA------A-ACAAA-AAUAA ........(((.(((((((..((.(((.((((((((.(((...))).))).))))).)))))....))))))).))).....------.-.....-..... ( -28.30) >DroEre_CAF1 17984 92 - 1 CAGGCGACUUGGUGCGCUUAAAGUCUUGGACUGACUGAAGCGACCUGCUGCCGCUUAAAGCCGCCAAAGCGCCGCAUAAAA-------A-ACAAG-CAAAC ..((((..((((((.((((..((((.......)))).(((((.........))))).))))))))))..))))((......-------.-....)-).... ( -30.50) >DroAna_CAF1 29522 92 - 1 CUGGCGACUUGGUGCGCUUAAAGUCUUGGACUGACUGAAGCGACCUGCAGCCGCUUAAAGCCGCCAAAGCGCCGCAGAAAA-------AAACAAACCAA-- ..((((..((((((.((((..((((.......)))).(((((.........))))).))))))))))..))))........-------...........-- ( -30.00) >DroPer_CAF1 26782 86 - 1 CAGGCAACUUGAGGCGCUUAAAGUCUUUGACUGACUGAAGCGACUGGGACCCGCUUGGAGCCGCCGAAGCGCAGCAUAAAA-------G-ACAA------- ..(((..(((.((((((((..((((.......)))).))))....((...)))))).)))..)))................-------.-....------- ( -21.70) >consensus CAGGCAACUUGGUGCGCUUAAAGUCUUCGACUGACUGAAGCGACCUGCAGCCGCUUAAAGCCGCCAAAGCGCCGCAUAAAA_______A_ACAAA_CAAAC ...((...((((((.((((..((((.......)))).(((((.........))))).)))))))))).))............................... (-17.60 = -17.80 + 0.20)

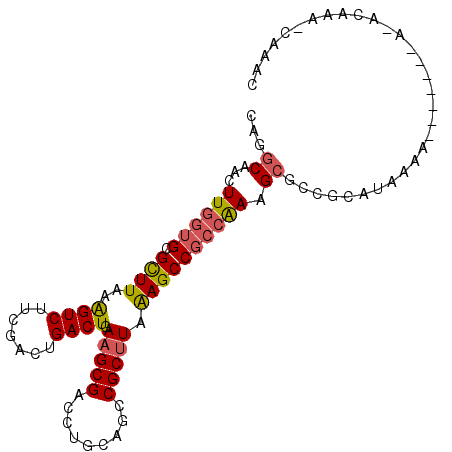

| Location | 18,114,151 – 18,114,252 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 77.60 |
| Mean single sequence MFE | -31.75 |
| Consensus MFE | -18.79 |
| Energy contribution | -19.68 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18114151 101 + 23771897 CUUUUGCGGCUUAAAGCGGCAGCAGGUCGCUUCAGUCAGUCGAAGACUUUAAGCGCACCAAGUCG------CCUGCCGCUCCACUCCAA-AAAAUACGAGACUUUCGG .....(((((.....(((((....((((((((.((((.......))))..))))).)))..))))------)..)))))..........-......((((...)))). ( -30.00) >DroVir_CAF1 18230 89 + 1 CUUUGGCGGCUUGAGGCGGCCCCAAGUCGCGUCAGUCAUUCGAAGACUUUAAACGCACCAAGUUG------CCUA--GUUCGGCUGCAC-G-A---CUACUC------ ....(((((((((..(((.....(((((...((........)).)))))....)))..)))))))------))((--((.((......)-)-)---)))...------ ( -30.10) >DroSim_CAF1 14884 100 + 1 CUUUUGCGGCUUAAAGCGGCAGCAGGUCGCUUCAGUCAGUCCAAGACUUUAAGCGCACCAAGUCG------CCUGCCGCUCCACUCCAA-AAA-GCCGAGUCUUUCGG (((((..((.....(((((((((.((((((((.((((.......))))..))))).))).....)------.)))))))).....)).)-)))-)(((((...))))) ( -32.50) >DroEre_CAF1 18007 100 + 1 CUUUGGCGGCUUUAAGCGGCAGCAGGUCGCUUCAGUCAGUCCAAGACUUUAAGCGCACCAAGUCG------CCUGCCGCUCCACUCGAA-AAG-GCCAAGACUUUCGG .((((((((.....(((((((((.((((((((.((((.......))))..))))).))).....)------.)))))))))).((....-.))-))))))........ ( -33.00) >DroYak_CAF1 18090 107 + 1 CUUUGGCGGCUUUAAGCGGCCGCAGGUCGCUUCAGUCAGUCCAAGACUUUAAGCGCACCAAGUCGCCUUCGCCUGCAGCUCCACUCAAAAAAA-GCCGAGACUUUCGG .((((((((((......))))((((((.((((.((((.......))))..))))((........))....)))))).................-))))))........ ( -31.30) >DroAna_CAF1 29545 78 + 1 CUUUGGCGGCUUUAAGCGGCUGCAGGUCGCUUCAGUCAGUCCAAGACUUUAAGCGCACCAAGUCG------CCAGCCGCUCCGU------------------------ ....(((((((....((((((...((((((((.((((.......))))..))))).))).)))))------).)))))))....------------------------ ( -33.60) >consensus CUUUGGCGGCUUUAAGCGGCAGCAGGUCGCUUCAGUCAGUCCAAGACUUUAAGCGCACCAAGUCG______CCUGCCGCUCCACUCCAA_AAA_GCCGAGACUUUCGG .....(((((......((((....((((((((.((((.......))))..))))).)))..)))).........)))))............................. (-18.79 = -19.68 + 0.89)


| Location | 18,114,151 – 18,114,252 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 77.60 |
| Mean single sequence MFE | -36.05 |
| Consensus MFE | -22.18 |
| Energy contribution | -23.42 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18114151 101 - 23771897 CCGAAAGUCUCGUAUUUU-UUGGAGUGGAGCGGCAGG------CGACUUGGUGCGCUUAAAGUCUUCGACUGACUGAAGCGACCUGCUGCCGCUUUAAGCCGCAAAAG .(((.....)))..((((-.(((..(((((((((((.------((....(((.(((((..((((.......)))).)))))))))))))))))))))..))).)))). ( -38.40) >DroVir_CAF1 18230 89 - 1 ------GAGUAG---U-C-GUGCAGCCGAAC--UAGG------CAACUUGGUGCGUUUAAAGUCUUCGAAUGACUGACGCGACUUGGGGCCGCCUCAAGCCGCCAAAG ------......---.-.-.....(((....--..))------)...((((((((((...((((.......)))))))))(.(((((((...))))))).)))))).. ( -28.00) >DroSim_CAF1 14884 100 - 1 CCGAAAGACUCGGC-UUU-UUGGAGUGGAGCGGCAGG------CGACUUGGUGCGCUUAAAGUCUUGGACUGACUGAAGCGACCUGCUGCCGCUUUAAGCCGCAAAAG ((((.....))))(-(((-((((..(((((((((((.------((....(((.(((((..((((.......)))).)))))))))))))))))))))..))).))))) ( -42.10) >DroEre_CAF1 18007 100 - 1 CCGAAAGUCUUGGC-CUU-UUCGAGUGGAGCGGCAGG------CGACUUGGUGCGCUUAAAGUCUUGGACUGACUGAAGCGACCUGCUGCCGCUUAAAGCCGCCAAAG .(....)..(((((-...-...(.((.(((((((((.------((....(((.(((((..((((.......)))).)))))))))))))))))))...)))))))).. ( -35.80) >DroYak_CAF1 18090 107 - 1 CCGAAAGUCUCGGC-UUUUUUUGAGUGGAGCUGCAGGCGAAGGCGACUUGGUGCGCUUAAAGUCUUGGACUGACUGAAGCGACCUGCGGCCGCUUAAAGCCGCCAAAG .(....)....(((-...(((((((((..(((((((((.(((....))).)..(((((..((((.......)))).))))).)))))))))))))))))..))).... ( -41.00) >DroAna_CAF1 29545 78 - 1 ------------------------ACGGAGCGGCUGG------CGACUUGGUGCGCUUAAAGUCUUGGACUGACUGAAGCGACCUGCAGCCGCUUAAAGCCGCCAAAG ------------------------...(.((((((((------((.((.(((.(((((..((((.......)))).))))))))...)).))))...))))))).... ( -31.00) >consensus CCGAAAGUCUCGGC_UUU_UUGGAGUGGAGCGGCAGG______CGACUUGGUGCGCUUAAAGUCUUGGACUGACUGAAGCGACCUGCUGCCGCUUAAAGCCGCCAAAG ........................((((((((((((((....)).....(((.(((((..((((.......)))).))))))))..)))))))).....))))..... (-22.18 = -23.42 + 1.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:15 2006