
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,092,561 – 18,092,741 |
| Length | 180 |
| Max. P | 0.845612 |

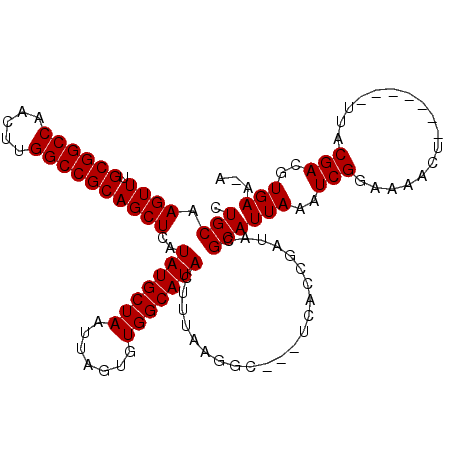
| Location | 18,092,561 – 18,092,669 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 90.65 |
| Mean single sequence MFE | -30.83 |
| Consensus MFE | -25.45 |
| Energy contribution | -25.45 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18092561 108 - 23771897 CUGCAAGUUUGCGGCCAACUUGGCCGCAGCUCAUAUGCUAAUUAGUGUGGCAUACUUUAAGGCCGCUCACCGAUACGCAUUAAAUCGCAAAACU-------UUACGACGUGAAAA .(((.((((.((((((.....))))))))))...((((..((..(((((((..........)))))..))..))..))))......)))....(-------((((...))))).. ( -28.90) >DroSec_CAF1 18937 107 - 1 CUGCAAGUUUGCGGCCAACUUGGCCGCAGCUCAUAUGCUAAUUAGUGUGGCAUACUUUAAGGCGGCCCACCGAUACGCAUUAAAUCGGAAAACU-------UUACGACGUGAA-A .(((......((((((.....))))))(((......))).((..(((.(((...(.....)...))))))..))..)))(((..(((.......-------...)))..))).-. ( -30.60) >DroEre_CAF1 19444 104 - 1 CUGCAAGUUUGCGGCCAACUUGGCCGCAGCUCAUAUGCUAAUUAGUGUGGCAUACUUUAAGGC---GCACGGCUAGGCAUUAAAUCGGAAAAGU-------UUACGACGUGAA-A ........((((((((.....)))))))).(((((((((.....(((((.(.........).)---)))).....)))))....(((.......-------...))).)))).-. ( -29.90) >DroYak_CAF1 26262 111 - 1 CUGCAAGUUUGCGGCCAACUUGGCCGCAGCUCAUAUGCUAAUUAGUGUGGCAUACUUUAAGGC---UCACCAAUAGGCAUUAAAUCGGAAAAGUUUCACGUUUACGACGUGAA-A (((..((((.((((((.....))))))))))...(((((.(((.(((.(((..........))---)))).))).))))).....))).....(((((((((...))))))))-) ( -33.90) >consensus CUGCAAGUUUGCGGCCAACUUGGCCGCAGCUCAUAUGCUAAUUAGUGUGGCAUACUUUAAGGC___UCACCGAUACGCAUUAAAUCGGAAAACU_______UUACGACGUGAA_A .(((.((((.((((((.....))))))))))..(((((((.......)))))))......................)))(((..(((.................)))..)))... (-25.45 = -25.45 + 0.00)


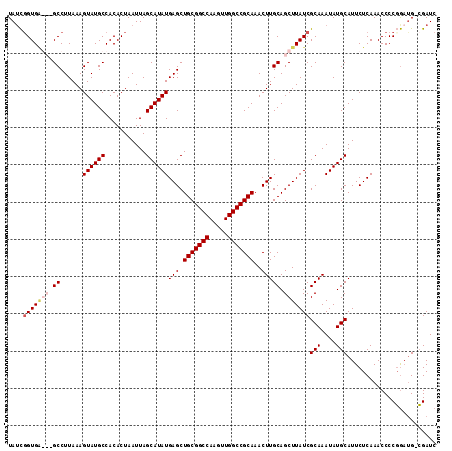
| Location | 18,092,594 – 18,092,708 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 93.42 |
| Mean single sequence MFE | -35.22 |
| Consensus MFE | -27.02 |
| Energy contribution | -28.40 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18092594 114 + 23771897 UAUCGGUGAGCGGCCUUAAAGUAUGCCACACUAAUUAGCAUAUGAGCUGCGGCCAAGUUGGCCGCAAACUUGCAGCUUAUCGCAAAUAUGCAUUCUCAAACCCCGGAUG-CGAUC ...((((((((.((......((((((...........))))))(((.(((((((.....)))))))..))))).))))))))......(((((((.........)))))-))... ( -38.20) >DroSec_CAF1 18969 114 + 1 UAUCGGUGGGCCGCCUUAAAGUAUGCCACACUAAUUAGCAUAUGAGCUGCGGCCAAGUUGGCCGCAAACUUGCAGCUUAUCGCAAAUAUGCAUUCUCAAACCCCGGAUG-CGAUC ...((((((((.((......((((((...........))))))(((.(((((((.....)))))))..))))).))))))))......(((((((.........)))))-))... ( -37.40) >DroEre_CAF1 19476 111 + 1 UAGCCGUGC---GCCUUAAAGUAUGCCACACUAAUUAGCAUAUGAGCUGCGGCCAAGUUGGCCGCAAACUUGCAGCUUAUCGCAAAUAUGCAUUCUCAAACCCCGGAUG-CGAUC ......(((---(.......((((((...........))))))(((((((((((.....))))(....)..)))))))..))))....(((((((.........)))))-))... ( -32.00) >DroYak_CAF1 26301 112 + 1 UAUUGGUGA---GCCUUAAAGUAUGCCACACUAAUUAGCAUAUGAGCUGCGGCCAAGUUGGCCGCAAACUUGCAGCUUAUCGCAAAUAUGCAUUCUCAAACCCCCACCGAGGAUC ....(((((---((......((((((...........))))))..(((((((((.....))))))).....)).)))))))(((....)))((((((...........)))))). ( -33.30) >consensus UAUCGGUGA___GCCUUAAAGUAUGCCACACUAAUUAGCAUAUGAGCUGCGGCCAAGUUGGCCGCAAACUUGCAGCUUAUCGCAAAUAUGCAUUCUCAAACCCCGGAUG_CGAUC ....(((((((.((......((((((...........))))))(((.(((((((.....)))))))..))))).)))))))(((....)))........................ (-27.02 = -28.40 + 1.38)


| Location | 18,092,594 – 18,092,708 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 93.42 |
| Mean single sequence MFE | -38.10 |
| Consensus MFE | -31.29 |
| Energy contribution | -32.73 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18092594 114 - 23771897 GAUCG-CAUCCGGGGUUUGAGAAUGCAUAUUUGCGAUAAGCUGCAAGUUUGCGGCCAACUUGGCCGCAGCUCAUAUGCUAAUUAGUGUGGCAUACUUUAAGGCCGCUCACCGAUA .....-....((((((.((((.......(((((((......)))))))((((((((.....))))))))))))...))).....(.(((((..........))))).).)))... ( -37.40) >DroSec_CAF1 18969 114 - 1 GAUCG-CAUCCGGGGUUUGAGAAUGCAUAUUUGCGAUAAGCUGCAAGUUUGCGGCCAACUUGGCCGCAGCUCAUAUGCUAAUUAGUGUGGCAUACUUUAAGGCGGCCCACCGAUA .....-....(((((........((((....))))....(((((.((((.((((((.....))))))))))..(((((((.......))))))).......))))))).)))... ( -38.70) >DroEre_CAF1 19476 111 - 1 GAUCG-CAUCCGGGGUUUGAGAAUGCAUAUUUGCGAUAAGCUGCAAGUUUGCGGCCAACUUGGCCGCAGCUCAUAUGCUAAUUAGUGUGGCAUACUUUAAGGC---GCACGGCUA .((((-((....(.(((....))).).....)))))).(((((((((((.((((((.....))))))))))....)))......(((((.(.........).)---)))))))). ( -37.30) >DroYak_CAF1 26301 112 - 1 GAUCCUCGGUGGGGGUUUGAGAAUGCAUAUUUGCGAUAAGCUGCAAGUUUGCGGCCAACUUGGCCGCAGCUCAUAUGCUAAUUAGUGUGGCAUACUUUAAGGC---UCACCAAUA .......((((((..((.(((.((((..(((((((......)))))))((((((((.....))))))))..(((((........))))))))).))).))..)---))))).... ( -39.00) >consensus GAUCG_CAUCCGGGGUUUGAGAAUGCAUAUUUGCGAUAAGCUGCAAGUUUGCGGCCAACUUGGCCGCAGCUCAUAUGCUAAUUAGUGUGGCAUACUUUAAGGC___UCACCGAUA .......(((((((((.((((.......(((((((......)))))))((((((((.....))))))))))))(((((((.......)))))))..........)))).))))). (-31.29 = -32.73 + 1.44)



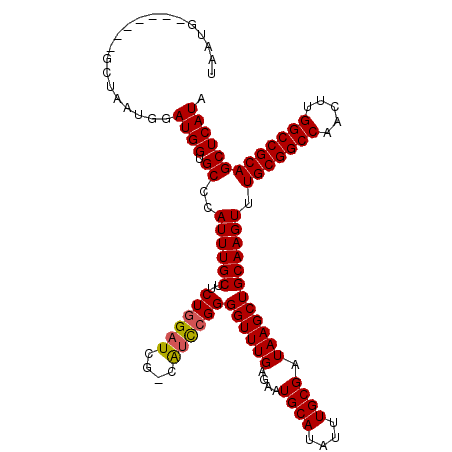
| Location | 18,092,634 – 18,092,741 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 93.24 |
| Mean single sequence MFE | -32.17 |
| Consensus MFE | -25.70 |
| Energy contribution | -26.95 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18092634 107 + 23771897 UAUGAGCUGCGGCCAAGUUGGCCGCAAACUUGCAGCUUAUCGCAAAUAUGCAUUCUCAAACCCCGGAUG-CGAUCCAGAAGCAAAUGGGCACCAUCCAUUAGC-------CAUUA .(((.(((((((((.....)))))).....(((.((.....)).....(((((((.........)))))-))........)))((((((.....)))))))))-------))).. ( -31.80) >DroSec_CAF1 19009 107 + 1 UAUGAGCUGCGGCCAAGUUGGCCGCAAACUUGCAGCUUAUCGCAAAUAUGCAUUCUCAAACCCCGGAUG-CGAUCCAGAAGCAAAUGGGCACCAUCCAUUAGC-------CAUUA .(((.(((((((((.....)))))).....(((.((.....)).....(((((((.........)))))-))........)))((((((.....)))))))))-------))).. ( -31.80) >DroEre_CAF1 19513 114 + 1 UAUGAGCUGCGGCCAAGUUGGCCGCAAACUUGCAGCUUAUCGCAAAUAUGCAUUCUCAAACCCCGGAUG-CGAUCCAGAAGCAAAUGGGCACCAUCCAUUAGCCAUUAGCCAUUA .(((((((((((((.....))))(....)..))))))))).((.....(((((((.........)))))-))........((.((((((.....)))))).)).....))..... ( -33.50) >DroYak_CAF1 26338 108 + 1 UAUGAGCUGCGGCCAAGUUGGCCGCAAACUUGCAGCUUAUCGCAAAUAUGCAUUCUCAAACCCCCACCGAGGAUCCAGCAGCAAAUUGGCACCAUCCAUUAGC-------CAUUA .....(((((((((.....))))(((.(.((((........)))).).)))((((((...........))))))...)))))....((((...........))-------))... ( -31.60) >consensus UAUGAGCUGCGGCCAAGUUGGCCGCAAACUUGCAGCUUAUCGCAAAUAUGCAUUCUCAAACCCCGGAUG_CGAUCCAGAAGCAAAUGGGCACCAUCCAUUAGC_______CAUUA .(((((((((((((.....))))(....)..))))))))).(((....))).............((((....))))....((.((((((.....)))))).))............ (-25.70 = -26.95 + 1.25)



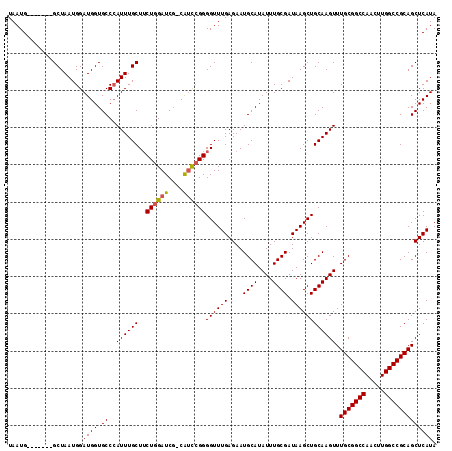
| Location | 18,092,634 – 18,092,741 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 93.24 |
| Mean single sequence MFE | -37.30 |
| Consensus MFE | -32.35 |
| Energy contribution | -32.48 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18092634 107 - 23771897 UAAUG-------GCUAAUGGAUGGUGCCCAUUUGCUUCUGGAUCG-CAUCCGGGGUUUGAGAAUGCAUAUUUGCGAUAAGCUGCAAGUUUGCGGCCAACUUGGCCGCAGCUCAUA ....(-------((.(((((.......))))).)))(((((((..-.)))))))...((((.......(((((((......)))))))((((((((.....)))))))))))).. ( -36.50) >DroSec_CAF1 19009 107 - 1 UAAUG-------GCUAAUGGAUGGUGCCCAUUUGCUUCUGGAUCG-CAUCCGGGGUUUGAGAAUGCAUAUUUGCGAUAAGCUGCAAGUUUGCGGCCAACUUGGCCGCAGCUCAUA ....(-------((.(((((.......))))).)))(((((((..-.)))))))...((((.......(((((((......)))))))((((((((.....)))))))))))).. ( -36.50) >DroEre_CAF1 19513 114 - 1 UAAUGGCUAAUGGCUAAUGGAUGGUGCCCAUUUGCUUCUGGAUCG-CAUCCGGGGUUUGAGAAUGCAUAUUUGCGAUAAGCUGCAAGUUUGCGGCCAACUUGGCCGCAGCUCAUA ...((((.....))))....((((.((..((((((..((..((((-((....(.(((....))).).....)))))).))..)))))).(((((((.....))))))))))))). ( -37.30) >DroYak_CAF1 26338 108 - 1 UAAUG-------GCUAAUGGAUGGUGCCAAUUUGCUGCUGGAUCCUCGGUGGGGGUUUGAGAAUGCAUAUUUGCGAUAAGCUGCAAGUUUGCGGCCAACUUGGCCGCAGCUCAUA ...((-------((...........))))....(((((.(((((((.....)))))))......(((.(((((((......))))))).)))((((.....)))))))))..... ( -38.90) >consensus UAAUG_______GCUAAUGGAUGGUGCCCAUUUGCUUCUGGAUCG_CAUCCGGGGUUUGAGAAUGCAUAUUUGCGAUAAGCUGCAAGUUUGCGGCCAACUUGGCCGCAGCUCAUA ....................((((.((..((((((..((((((....))))))((((((....((((....)))).)))))))))))).(((((((.....))))))))))))). (-32.35 = -32.48 + 0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:07 2006