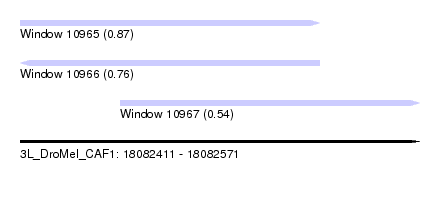
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,082,411 – 18,082,571 |
| Length | 160 |
| Max. P | 0.873856 |

| Location | 18,082,411 – 18,082,531 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.81 |
| Mean single sequence MFE | -41.73 |
| Consensus MFE | -38.75 |
| Energy contribution | -39.75 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
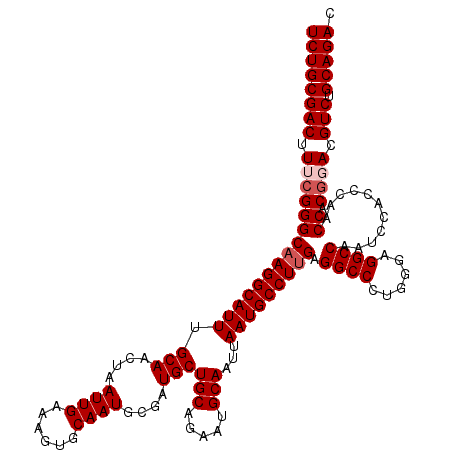
>3L_DroMel_CAF1 18082411 120 + 23771897 UCUGCGACUUUCGGGCAAGGCAUUUGCAACUAAUUGAAAGUGCAAUGCGAUGCUGCAGAAUGCAAUUAAUGCCUUGAGGCCCUGGGAGGCCAAUCCACCCAACCACGGACGUCUGCAGAC ((((((((.(((((((((((((((.(((....((((......))))....)))(((.....)))...))))))))).((((......))))...........)).)))).))).))))). ( -42.50) >DroSec_CAF1 8677 120 + 1 UCUGCGACUUUCGGGCAAGGCAUUUGCAACUAAUUGAAAGUGCAAUGCGAUGCUGCAGAAUGCAAUUAAUGCCUUGAGGCCCUGGGAGGCCAAUCCACCCAACCACGGACGUCUGCAGAC ((((((((.(((((((((((((((.(((....((((......))))....)))(((.....)))...))))))))).((((......))))...........)).)))).))).))))). ( -42.50) >DroEre_CAF1 9420 120 + 1 UCUGCGACUUUGGGGCAAGGCAUUUGCAACUAAUUGAAAGUGCAAUGCGAUGCUGCAGAAUGCAAUUAAUGCCUUGAGGCCCUGGGAGGCCAAUCCACCCAACCACGUACGUCUGCAGAC ((((((((...(((.(((((((((.(((....((((......))))....)))(((.....)))...)))))))))...)))((((.((.....)).)))).........))).))))). ( -40.50) >DroYak_CAF1 11738 120 + 1 UCUGCGACUUUCGGGCAAGGCAUUUGCAACUAAUUGAAAGUGCAAUGCGAUGCUGCAGAAUGCAAUUAAUGCCUCGAGGCCCUGGGAGGCCAGUACUCCCCACCACGCACGUCUGCAGAC ((((((((....((((.(((((((.(((....((((......))))....)))(((.....)))...)))))))....)))).(((((.......)))))..........))).))))). ( -41.40) >consensus UCUGCGACUUUCGGGCAAGGCAUUUGCAACUAAUUGAAAGUGCAAUGCGAUGCUGCAGAAUGCAAUUAAUGCCUUGAGGCCCUGGGAGGCCAAUCCACCCAACCACGGACGUCUGCAGAC ((((((((.(((((((((((((((.(((....((((......))))....)))(((.....)))...))))))))).((((......))))...........)).)))).))).))))). (-38.75 = -39.75 + 1.00)

| Location | 18,082,411 – 18,082,531 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.81 |
| Mean single sequence MFE | -42.88 |
| Consensus MFE | -40.29 |
| Energy contribution | -40.35 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.758547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18082411 120 - 23771897 GUCUGCAGACGUCCGUGGUUGGGUGGAUUGGCCUCCCAGGGCCUCAAGGCAUUAAUUGCAUUCUGCAGCAUCGCAUUGCACUUUCAAUUAGUUGCAAAUGCCUUGCCCGAAAGUCGCAGA .(((((.(((.........((((.((.....)).))))((((...((((((((..((((.....))))........((((((.......)).))))))))))))))))....)))))))) ( -42.70) >DroSec_CAF1 8677 120 - 1 GUCUGCAGACGUCCGUGGUUGGGUGGAUUGGCCUCCCAGGGCCUCAAGGCAUUAAUUGCAUUCUGCAGCAUCGCAUUGCACUUUCAAUUAGUUGCAAAUGCCUUGCCCGAAAGUCGCAGA .(((((.(((.........((((.((.....)).))))((((...((((((((..((((.....))))........((((((.......)).))))))))))))))))....)))))))) ( -42.70) >DroEre_CAF1 9420 120 - 1 GUCUGCAGACGUACGUGGUUGGGUGGAUUGGCCUCCCAGGGCCUCAAGGCAUUAAUUGCAUUCUGCAGCAUCGCAUUGCACUUUCAAUUAGUUGCAAAUGCCUUGCCCCAAAGUCGCAGA .(((((.(((.........((((.((.....)).))))(((...(((((((((..((((.....))))........((((((.......)).))))))))))))).)))...)))))))) ( -42.40) >DroYak_CAF1 11738 120 - 1 GUCUGCAGACGUGCGUGGUGGGGAGUACUGGCCUCCCAGGGCCUCGAGGCAUUAAUUGCAUUCUGCAGCAUCGCAUUGCACUUUCAAUUAGUUGCAAAUGCCUUGCCCGAAAGUCGCAGA .(((((.(((...((.(((((((....((((....))))..))))((((((((..((((.....))))........((((((.......)).)))))))))))))))))...)))))))) ( -43.70) >consensus GUCUGCAGACGUCCGUGGUUGGGUGGAUUGGCCUCCCAGGGCCUCAAGGCAUUAAUUGCAUUCUGCAGCAUCGCAUUGCACUUUCAAUUAGUUGCAAAUGCCUUGCCCGAAAGUCGCAGA .(((((.(((..........(((.((.....)).))).((((...((((((((..((((.....))))........((((((.......)).))))))))))))))))....)))))))) (-40.29 = -40.35 + 0.06)

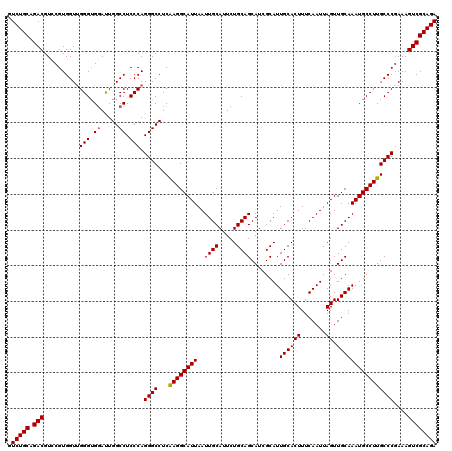
| Location | 18,082,451 – 18,082,571 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.81 |
| Mean single sequence MFE | -34.88 |
| Consensus MFE | -31.30 |
| Energy contribution | -31.80 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
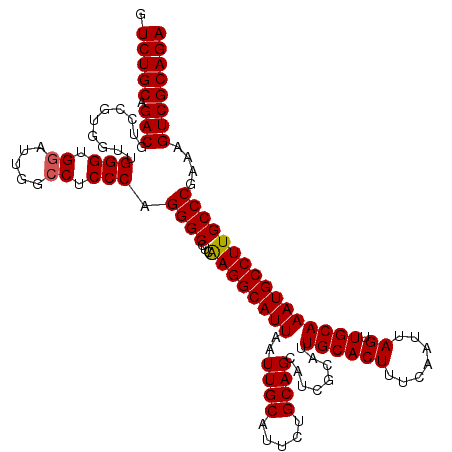
>3L_DroMel_CAF1 18082451 120 + 23771897 UGCAAUGCGAUGCUGCAGAAUGCAAUUAAUGCCUUGAGGCCCUGGGAGGCCAAUCCACCCAACCACGGACGUCUGCAGACGAGAAGUUGCAUUCGAAACCGAAAUGCACAGAAAAAACGA ((((.((((....))))((((((((((...(((....)))..((((.((.....)).))))........((((....))))...))))))))))..........))))............ ( -33.50) >DroSec_CAF1 8717 120 + 1 UGCAAUGCGAUGCUGCAGAAUGCAAUUAAUGCCUUGAGGCCCUGGGAGGCCAAUCCACCCAACCACGGACGUCUGCAGACGAGAAGUUGCAUUCGAAACCGAAAUGCACAGAAAAAACGA ((((.((((....))))((((((((((...(((....)))..((((.((.....)).))))........((((....))))...))))))))))..........))))............ ( -33.50) >DroEre_CAF1 9460 120 + 1 UGCAAUGCGAUGCUGCAGAAUGCAAUUAAUGCCUUGAGGCCCUGGGAGGCCAAUCCACCCAACCACGUACGUCUGCAGACGAGAAGUUGCAUUCGAAACCGAAAUGCACAGAAAAAACGA ((((.((((((.(((((((.(((.......(((....)))..((((.((.....)).)))).....)))..))))))).......))))))((((....)))).))))............ ( -34.50) >DroYak_CAF1 11778 120 + 1 UGCAAUGCGAUGCUGCAGAAUGCAAUUAAUGCCUCGAGGCCCUGGGAGGCCAGUACUCCCCACCACGCACGUCUGCAGACGAGAAGUUGCAUUCGAAACCGAAAUGCACACAAAAAACGA ((((.((((((.(((((((.(((.......(((....)))...(((((.......)))))......)))..))))))).......))))))((((....)))).))))............ ( -38.00) >consensus UGCAAUGCGAUGCUGCAGAAUGCAAUUAAUGCCUUGAGGCCCUGGGAGGCCAAUCCACCCAACCACGGACGUCUGCAGACGAGAAGUUGCAUUCGAAACCGAAAUGCACAGAAAAAACGA ((((.((((....))))((((((((((...(((....)))..((((.((.....)).))))........((((....))))...))))))))))..........))))............ (-31.30 = -31.80 + 0.50)

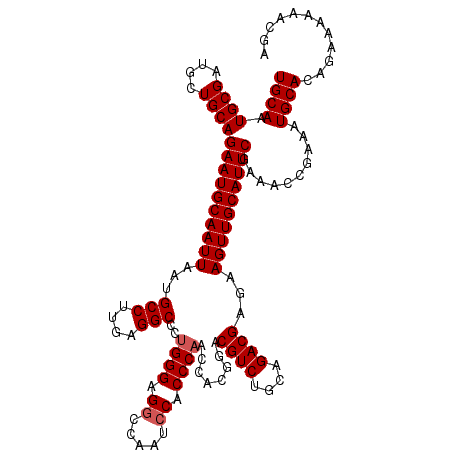
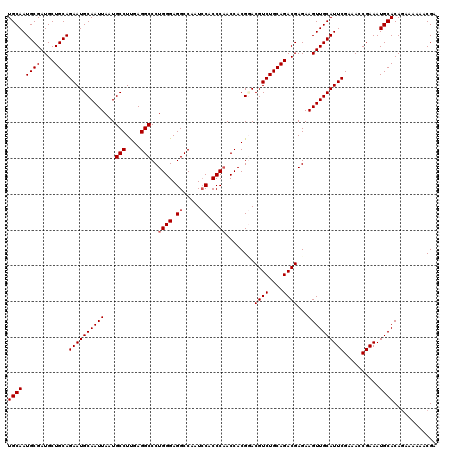
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:58 2006