| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
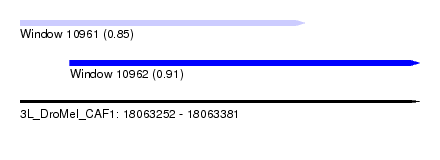
| Location | 18,063,252 – 18,063,381 |
| Length | 129 |
| Max. P | 0.911057 |

| Location | 18,063,252 – 18,063,344 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 80.50 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -18.26 |
| Energy contribution | -19.24 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18063252 92 + 23771897 AAAAGUGCCCAACGAUGCAGUCGAUAAAUCACGUGCGCUGCUCAUCUCAAAAGGCGAUUGGGAUUGCGAUUGUGUCAGCCG--UG-GCAAUGCAA .....(((.....((((((((((...((((.....((((.............)))).....)))).)))))))))).((..--..-))...))). ( -26.52) >DroSec_CAF1 8841 92 + 1 AAAAGUGCCAAGCGAUGCAGUCGAUAAAUCACGUGCGCUGCUCAUCUCAAAAGGCGAUUGGGAUUGCGAUUGUGUCAGCCG--UG-GCAAUGCAA .....(((((.((((((((((((...((((.....((((.............)))).....)))).)))))))))).))..--))-)))...... ( -30.72) >DroSim_CAF1 9175 92 + 1 AAAAGUGCCCAGCGAUGCAGUCGAUAAAUCACGUGCGCUGCUCAUCUCAAAAGGCGAUUGUGAUUGUGAUUGUGUCAGCCG--UG-GCAAUGCAA .....(((((.((((((((((((...(((((((..((((.............))))..))))))).)))))))))).)).)--.)-)))...... ( -32.92) >DroEre_CAF1 8787 92 + 1 AAAAGUGCCCAGCGAUGCAGUCGAUAAAUCACGUGCGCUGCUCAUCUCAAUGGGCGAUUGUGAUUGCGAUUGUGUCAGCCG--UG-GCAAUGCGA .....(((((.((((((((((((...(((((((.....(((((((....)))))))..))))))).)))))))))).)).)--.)-)))...... ( -36.00) >DroYak_CAF1 9655 95 + 1 AAAAGUGCCCAGCGAUGCAGUCGAUAAAUCACGUGCGCUGCUCAUCUCAAAAGGCGACUGUGAUUGUGAUUGCGAUUGCAAAAUGCAAAAUGCAA ...........((((((((((((...(((((((..((((.............))))..))))))).))))))).)))))....(((.....))). ( -29.72) >DroAna_CAF1 7556 73 + 1 CUGAGUCUC---CGAUGCAGUCGAUAAAUCACGUUCGGCACUCAUCUCAGAGUGCGAUUAUGAUUAAAAUUCUGUU------------------- .........---....((((......(((((...(((.(((((......))))))))...)))))......)))).------------------- ( -16.90) >consensus AAAAGUGCCCAGCGAUGCAGUCGAUAAAUCACGUGCGCUGCUCAUCUCAAAAGGCGAUUGUGAUUGCGAUUGUGUCAGCCG__UG_GCAAUGCAA .............((((((((((...(((((((..((((.............))))..))))))).))))))))))................... (-18.26 = -19.24 + 0.97)



| Location | 18,063,268 – 18,063,381 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 89.51 |
| Mean single sequence MFE | -38.62 |
| Consensus MFE | -28.00 |
| Energy contribution | -28.16 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.911057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18063268 113 + 23771897 GCAGUCGAUAAAUCACGUGCGCUGCUCAUCUCAAAAGGCGAUUGGGAUUGCGAUUGUGUCAGCCG--UG-GCAAUGCAAAAUUAAAAGGAGCUGUUGCCAUGGCUAAGACUUU-UUG (((((((...((((.....((((.............)))).....)))).)))))))((((((((--((-(((((((.............)).))))))))))))..)))...-... ( -39.44) >DroSec_CAF1 8857 113 + 1 GCAGUCGAUAAAUCACGUGCGCUGCUCAUCUCAAAAGGCGAUUGGGAUUGCGAUUGUGUCAGCCG--UG-GCAAUGCAAAAUGAAAAGGAGCUGUUGCCAUGGCUCAGACUUU-UUG (((((((...((((.....((((.............)))).....)))).)))))))((((((((--((-(((((((....(....)...)).))))))))))))..)))...-... ( -39.72) >DroSim_CAF1 9191 113 + 1 GCAGUCGAUAAAUCACGUGCGCUGCUCAUCUCAAAAGGCGAUUGUGAUUGUGAUUGUGUCAGCCG--UG-GCAAUGCAAAAUUAAAAGGAGCUGUUGCCAUGGCUCAGACUUU-UUG (((((((...(((((((..((((.............))))..))))))).)))))))((((((((--((-(((((((.............)).))))))))))))..)))...-... ( -42.24) >DroEre_CAF1 8803 111 + 1 GCAGUCGAUAAAUCACGUGCGCUGCUCAUCUCAAUGGGCGAUUGUGAUUGCGAUUGUGUCAGCCG--UG-GCAAUGCGAAAUUGAAAU--GUUGUUGCCAUGGCUCAGACUUU-UUG (((((((...(((((((.....(((((((....)))))))..))))))).)))))))((((((((--((-(((((.............--...))))))))))))..)))...-... ( -43.29) >DroYak_CAF1 9671 115 + 1 GCAGUCGAUAAAUCACGUGCGCUGCUCAUCUCAAAAGGCGACUGUGAUUGUGAUUGCGAUUGCAAAAUGCAAAAUGCAAAAUGAAAAU--GCUGUUUCCAGGGCUCAGACUUUUUUG (((((((...(((((((..(((.(.((..((....))..))).)))..))))))).)))))))....(((.....)))....(((((.--.(((...(....)..)))...))))). ( -28.40) >consensus GCAGUCGAUAAAUCACGUGCGCUGCUCAUCUCAAAAGGCGAUUGUGAUUGCGAUUGUGUCAGCCG__UG_GCAAUGCAAAAUUAAAAGGAGCUGUUGCCAUGGCUCAGACUUU_UUG (((((((...(((((((..((((.............))))..))))))).)))))))(((((((...((.(((((((.............)).))))))).))))..)))....... (-28.00 = -28.16 + 0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:54 2006