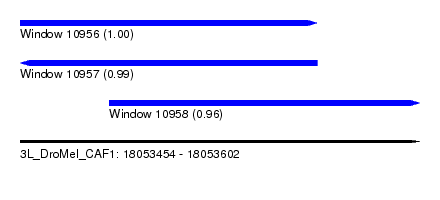
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,053,454 – 18,053,602 |
| Length | 148 |
| Max. P | 0.997672 |

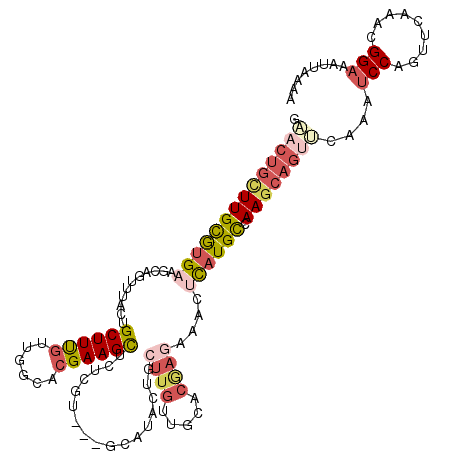
| Location | 18,053,454 – 18,053,564 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.50 |
| Mean single sequence MFE | -34.13 |
| Consensus MFE | -13.72 |
| Energy contribution | -15.64 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.40 |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.997672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18053454 110 + 23771897 -------UUGCGUGAAGAUGUUAACUGCUUUAUUCGCACGAAGCUCAUGU---GCAUACUGCUUGUUGCACGAAAAACGCAUGCCAAGCAGUUCAAAUCCAGUUCAAACGGAAAUUAGUA -------.(((.......(((((((((........(((((.......)))---))..((((((((.(((.........)))...)))))))).......)))))..)))).......))) ( -23.54) >DroSec_CAF1 168120 117 + 1 CAACUGCUUGCAUGAAGUCGUUUACUGCUUUGUUGGUGCGAAGUUUUCGU---GUGGACUGCUUGUUGCACGAGAAACUCAUGCCAAGCAGUUCAAAUCCAGUUCAAACGGAAAUGAAAA ..(((((((((((((.((........(((((((....)))))))((((((---(..(.(.....))..))))))).))))))).))))))))(((..(((.((....)))))..)))... ( -37.50) >DroSim_CAF1 179965 117 + 1 GAACUGUUUGCAUGAAGUCGUUUACUGCUUUGUAGUCACGAAGUUUUCGU---GCAUACUGCUUGUUGCACGAGAAACUCAUGCCAAGCAGUUCAAAUCCAGUUCAAACGGAAAUGAAAA (((((((((((((((..((((..(((((...))))).))))..(((((((---(((.((.....))))))))))))..))))).))))))))))...(((.((....)))))........ ( -41.90) >DroEre_CAF1 168204 109 + 1 GUACUGCUUGUGCGAAACAGUUUACUGCUUUGUUGGCACGAAGC-----------ACACUGCUUGUAGGACAAGAAACUCAUGUAAAGCAGUACAAAACCAGUUCAAACGGAAACUAAAC (((((((((.((((...((((....((((((((....)))))))-----------).))))((((.....)))).......)))))))))))))...............(....)..... ( -30.50) >DroYak_CAF1 175942 110 + 1 UAACUGCUUGUGUGGUACAGUUUACUGCUUCGCUGGCACGAAGCUCUUGUUAAGCGUACUGCUUGUGG-------AACGUAUGUAAAGCAGG--AAAUCCACUUCAAACGGAAG-UGGAU ...((((((..(..((((.((((((.((((((......))))))....).)))))))))..)......-------.((....)).)))))).--..(((((((((.....))))-))))) ( -37.20) >consensus GAACUGCUUGCGUGAAGCAGUUUACUGCUUUGUUGGCACGAAGCUCUCGU___GCAUACUGCUUGUUGCACGAGAAACUCAUGCCAAGCAGUUCAAAUCCAGUUCAAACGGAAAUUAAAA .((((((((((((((...........((((((......)))))).................((((.....))))....)))))).))))))))....(((.........)))........ (-13.72 = -15.64 + 1.92)

| Location | 18,053,454 – 18,053,564 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.50 |
| Mean single sequence MFE | -31.66 |
| Consensus MFE | -15.82 |
| Energy contribution | -19.42 |
| Covariance contribution | 3.60 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.50 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18053454 110 - 23771897 UACUAAUUUCCGUUUGAACUGGAUUUGAACUGCUUGGCAUGCGUUUUUCGUGCAACAAGCAGUAUGC---ACAUGAGCUUCGUGCGAAUAAAGCAGUUAACAUCUUCACGCAA------- ...........((.(((...((((...((((((((.(((((.((((...(((((((.....)).)))---))..))))..))))).....))))))))...))))))).))..------- ( -29.70) >DroSec_CAF1 168120 117 - 1 UUUUCAUUUCCGUUUGAACUGGAUUUGAACUGCUUGGCAUGAGUUUCUCGUGCAACAAGCAGUCCAC---ACGAAAACUUCGCACCAACAAAGCAGUAAACGACUUCAUGCAAGCAGUUG ...(((..((((.......))))..)))((((((((.(((((((((.(((((..((.....))...)---))))))))...((.........))...........))))))))))))).. ( -31.10) >DroSim_CAF1 179965 117 - 1 UUUUCAUUUCCGUUUGAACUGGAUUUGAACUGCUUGGCAUGAGUUUCUCGUGCAACAAGCAGUAUGC---ACGAAAACUUCGUGACUACAAAGCAGUAAACGACUUCAUGCAAACAGUUC ...............((((((.......(((((((((((((((...)))))))..)))))))).(((---(.(((....((((...(((......))).)))).))).))))..)))))) ( -35.90) >DroEre_CAF1 168204 109 - 1 GUUUAGUUUCCGUUUGAACUGGUUUUGUACUGCUUUACAUGAGUUUCUUGUCCUACAAGCAGUGU-----------GCUUCGUGCCAACAAAGCAGUAAACUGUUUCGCACAAGCAGUAC ..(((((((......)))))))....((((((((((((((......((((.....))))..))))-----------)....((((.....((((((....)))))).))))))))))))) ( -28.40) >DroYak_CAF1 175942 110 - 1 AUCCA-CUUCCGUUUGAAGUGGAUUU--CCUGCUUUACAUACGUU-------CCACAAGCAGUACGCUUAACAAGAGCUUCGUGCCAGCGAAGCAGUAAACUGUACCACACAAGCAGUUA (((((-((((.....)))))))))..--.(((((((((((((...-------....((((.....)))).......(((((((....))))))).)))...))))......))))))... ( -33.20) >consensus UUUUAAUUUCCGUUUGAACUGGAUUUGAACUGCUUGGCAUGAGUUUCUCGUGCAACAAGCAGUAUGC___ACAAAAGCUUCGUGCCAACAAAGCAGUAAACGACUUCACGCAAGCAGUUC ...........(((((...((((.(((.(((((((((((((((...)))))))..)))))))).............((((.((....)).))))......))).))))..)))))..... (-15.82 = -19.42 + 3.60)

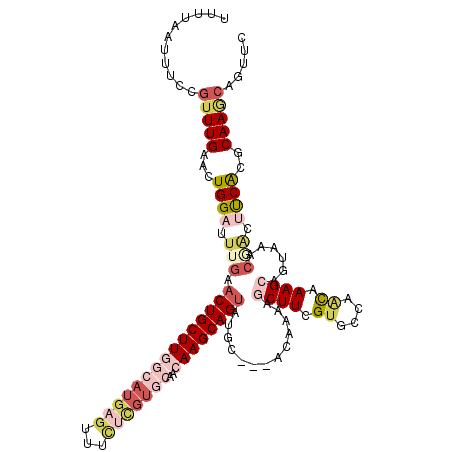
| Location | 18,053,487 – 18,053,602 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.39 |
| Mean single sequence MFE | -23.52 |
| Consensus MFE | -7.12 |
| Energy contribution | -9.08 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.30 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18053487 115 + 23771897 AAGCUCAUGU---GCAUACUGCUUGUUGCACGAAAAACGCAUGCCAAGCAGUUCAAAUCCAGUUCAAACGGAAAUUAGUAAUCAUGC-AAUCCUGC-AUUUAUUUAUUUGUAUAUCAUUA ..((.....(---((((((((((((.(((.........)))...)))))))).....(((.((....)))))...........))))-).....))-....................... ( -22.90) >DroSec_CAF1 168160 116 + 1 AAGUUUUCGU---GUGGACUGCUUGUUGCACGAGAAACUCAUGCCAAGCAGUUCAAAUCCAGUUCAAACGGAAAUGAAAAAUCAUGCCAAUCCUGC-AUUUAUUUAUUUUUAUAACAUUA ...(((((((---.(((((((((((..(((.(((...))).))))))))))))))..(((.((....))))).)))))))...((((.......))-))..................... ( -28.20) >DroSim_CAF1 180005 113 + 1 AAGUUUUCGU---GCAUACUGCUUGUUGCACGAGAAACUCAUGCCAAGCAGUUCAAAUCCAGUUCAAACGGAAAUGAAAAAUCAUGC-AAUC--GC-AUUUAUUUAUUUUUAUAACAUUA ..((.....(---((((.(((((((..(((.(((...))).))))))))))((((..(((.((....)))))..)))).....))))-)...--))-....................... ( -23.90) >DroEre_CAF1 168244 103 + 1 AAGC-----------ACACUGCUUGUAGGACAAGAAACUCAUGUAAAGCAGUACAAAACCAGUUCAAACGGAAACUAAACAU------AAUUUCACUAAACAUUUAUUUCUAUAAUAUUC ....-----------..(((((((.....(((.(.....).))).))))))).................(....).......------................................ ( -11.10) >DroYak_CAF1 175982 103 + 1 AAGCUCUUGUUAAGCGUACUGCUUGUGG-------AACGUAUGUAAAGCAGG--AAAUCCACUUCAAACGGAAG-UGGAUGC------AAUCUUGC-AUUUAUUUAUUUCUAUAAUAUUU ..(((.......))).......((((((-------((.(((.((((((((((--(.(((((((((.....))))-)))))..------..))))))-.))))).)))))))))))..... ( -31.50) >consensus AAGCUCUCGU___GCAUACUGCUUGUUGCACGAGAAACUCAUGCCAAGCAGUUCAAAUCCAGUUCAAACGGAAAUUAAAAAUCAUGC_AAUCCUGC_AUUUAUUUAUUUCUAUAACAUUA .................((((((((..(((.((.....)).))))))))))).....(((.........)))................................................ ( -7.12 = -9.08 + 1.96)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:50 2006