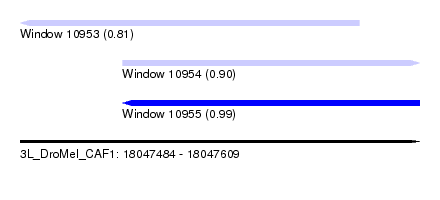
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 18,047,484 – 18,047,609 |
| Length | 125 |
| Max. P | 0.987507 |

| Location | 18,047,484 – 18,047,590 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 81.48 |
| Mean single sequence MFE | -35.74 |
| Consensus MFE | -24.75 |
| Energy contribution | -25.76 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18047484 106 - 23771897 UUCUCUCUGGCUGGGAAUCUUCUCCCCUCAAACU-CCCAAAGCCAGGGGCCUAUC-----GCUCGGGCUAAUGGCCUCUCGAGUCGCUAGCCGAUGCUAUAAUCGUCGGAGU ...(((((((((((((.................)-)))..))))))))).(((.(-----((((((((.....)))...)))).)).)))((((((.......))))))... ( -35.73) >DroPse_CAF1 162758 95 - 1 UC---------UGUCCCUCCUCUCCCC----ACUGCCCCCACCCA----CCCAUCAAUAGGCAUAGGCUAAUGGCCUCUCGAGUCGUUCGCUGAUGCUAUAAUCGUUGGGAU ..---------................----..............----((((......(((((.(((.(((((((....).)))))).))).)))))........)))).. ( -20.14) >DroSec_CAF1 162345 106 - 1 UUCUCUCUGGCUGGGAAUCUUCUCCCCUUAAACU-CCCAAAGCCAGGGGCCUAAC-----GCUCGGGCUAAUGGCCUCUCGAGUCGCCAGCCGAUGCUAUAAUCGUCGGAUU ......(((((.((((......)))).....(((-(.....((((..(((((...-----....)))))..)))).....)))).)))))((((((.......))))))... ( -37.70) >DroSim_CAF1 174169 106 - 1 UUCUCUCUGGCUGGGAAUCUUCUCCCCUUAAACU-CCCAAAGCCAGGGGCCUAUC-----GCUCGGGCUAAUGGCCUUUCGAGUCGCCAGCCGAUGCUAUAAUCGUCGGAGU ......(((((.((((......)))).....(((-(..((.((((..(((((...-----....)))))..)))).))..)))).)))))((((((.......))))))... ( -37.80) >DroEre_CAF1 162622 106 - 1 CUCUCUCUGGCUGGGAAUCUUCUCCCCUGGAACU-CCCAAAGCCAGGAGCCUAUC-----GCUCCGGCUAAUGGCCUCUCCAGUCGCCAGCCGACGCGAUGGUCGUCGGAGU ......(((((.((((......))))(((((...-.(((.((((.(((((.....-----)))))))))..)))....)))))..)))))(((((((....).))))))... ( -46.00) >DroYak_CAF1 170247 106 - 1 UUCUCUCUGGCUGGGAAUCUUCUCCCCUUGAACU-CCCAGAGCCAGGAGCCUAUC-----GCUCGGGCUAAUGGCCUCUCCAGUCGCUAGCCGACGCCAUAAUCGUCGGAGU ......((((((((((...(((.......))).)-))))).((((..(((((...-----....)))))..))))....)))).......((((((.......))))))... ( -37.10) >consensus UUCUCUCUGGCUGGGAAUCUUCUCCCCUUAAACU_CCCAAAGCCAGGGGCCUAUC_____GCUCGGGCUAAUGGCCUCUCGAGUCGCCAGCCGAUGCUAUAAUCGUCGGAGU ......(((((.((((......))))...............((((..((((..............))))..))))..........)))))((((((.......))))))... (-24.75 = -25.76 + 1.00)


| Location | 18,047,516 – 18,047,609 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 94.09 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -29.90 |
| Energy contribution | -29.70 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18047516 93 + 23771897 GAGAGGCCAUUAGCCCGAGCGAUAGGCCCCUGGCUUUGGGAGUUUGAGGGGAGAAGAUUCCCAGCCAGAGAGAACUCUACGGUAACUCUUCCU (((.((((....((....))....)))).((((((..((((((((.........))))))))))))))......)))...((........)). ( -31.00) >DroSec_CAF1 162377 93 + 1 GAGAGGCCAUUAGCCCGAGCGUUAGGCCCCUGGCUUUGGGAGUUUAAGGGGAGAAGAUUCCCAGCCAGAGAGAACUCUACGGUAACUCUUUCU ....(((......((((((.(((((....))))))))))).......(((((.....))))).)))(((((((...(....)....))))))) ( -31.40) >DroSim_CAF1 174201 93 + 1 GAAAGGCCAUUAGCCCGAGCGAUAGGCCCCUGGCUUUGGGAGUUUAAGGGGAGAAGAUUCCCAGCCAGAGAGAACUCUACGGUAACUCUUUCU ((((((((....((....))....)))).((((((..((((((((.........)))))))))))))).(((....(....)...))))))). ( -30.10) >DroEre_CAF1 162654 93 + 1 GAGAGGCCAUUAGCCGGAGCGAUAGGCUCCUGGCUUUGGGAGUUCCAGGGGAGAAGAUUCCCAGCCAGAGAGAGCUCUACGGUAACUCUCCCU ...((((((..(((((((((.....))))).)))).)))(((((((.(((((.....)))))....((((....))))..)).)))))..))) ( -37.90) >DroYak_CAF1 170279 93 + 1 GAGAGGCCAUUAGCCCGAGCGAUAGGCUCCUGGCUCUGGGAGUUCAAGGGGAGAAGAUUCCCAGCCAGAGAGAACUCUACGGUAACUCUCUCG ((((..(((..((((.((((.....))))..)))).)))(((((...(((((.....))))).(((((((....))))..)))))))))))). ( -34.00) >consensus GAGAGGCCAUUAGCCCGAGCGAUAGGCCCCUGGCUUUGGGAGUUUAAGGGGAGAAGAUUCCCAGCCAGAGAGAACUCUACGGUAACUCUUUCU ..(((((((...(((.........)))...)))))))(((((((...(((((.....))))).(((((((....))))..))))))))))... (-29.90 = -29.70 + -0.20)


| Location | 18,047,516 – 18,047,609 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 94.09 |
| Mean single sequence MFE | -31.86 |
| Consensus MFE | -27.82 |
| Energy contribution | -27.78 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 18047516 93 - 23771897 AGGAAGAGUUACCGUAGAGUUCUCUCUGGCUGGGAAUCUUCUCCCCUCAAACUCCCAAAGCCAGGGGCCUAUCGCUCGGGCUAAUGGCCUCUC .((..(((((....(((((....)))))...((((......))))....)))))))...((((..(((((.......)))))..))))..... ( -29.40) >DroSec_CAF1 162377 93 - 1 AGAAAGAGUUACCGUAGAGUUCUCUCUGGCUGGGAAUCUUCUCCCCUUAAACUCCCAAAGCCAGGGGCCUAACGCUCGGGCUAAUGGCCUCUC .....(((((....(((((....)))))...((((......))))....))))).....((((..(((((.......)))))..))))..... ( -28.00) >DroSim_CAF1 174201 93 - 1 AGAAAGAGUUACCGUAGAGUUCUCUCUGGCUGGGAAUCUUCUCCCCUUAAACUCCCAAAGCCAGGGGCCUAUCGCUCGGGCUAAUGGCCUUUC .(((((((((....(((((....)))))...((((......))))....))))).....((((..(((((.......)))))..)))).)))) ( -28.20) >DroEre_CAF1 162654 93 - 1 AGGGAGAGUUACCGUAGAGCUCUCUCUGGCUGGGAAUCUUCUCCCCUGGAACUCCCAAAGCCAGGAGCCUAUCGCUCCGGCUAAUGGCCUCUC ((((((((((.......))))))))))((((((((.(((........)))..))))..((((.(((((.....)))))))))...)))).... ( -39.60) >DroYak_CAF1 170279 93 - 1 CGAGAGAGUUACCGUAGAGUUCUCUCUGGCUGGGAAUCUUCUCCCCUUGAACUCCCAGAGCCAGGAGCCUAUCGCUCGGGCUAAUGGCCUCUC .(((((........(((((....))))).((((((...(((.......))).)))))).((((..(((((.......)))))..))))))))) ( -34.10) >consensus AGAAAGAGUUACCGUAGAGUUCUCUCUGGCUGGGAAUCUUCUCCCCUUAAACUCCCAAAGCCAGGGGCCUAUCGCUCGGGCUAAUGGCCUCUC .(((((((((.(((.(((....))).)))..((((......))))....))))).....((((..(((((.......)))))..)))).)))) (-27.82 = -27.78 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:47 2006