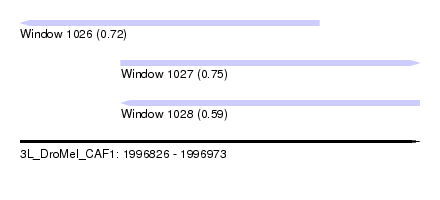
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,996,826 – 1,996,973 |
| Length | 147 |
| Max. P | 0.745335 |

| Location | 1,996,826 – 1,996,936 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 84.20 |
| Mean single sequence MFE | -31.24 |
| Consensus MFE | -17.84 |
| Energy contribution | -19.44 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
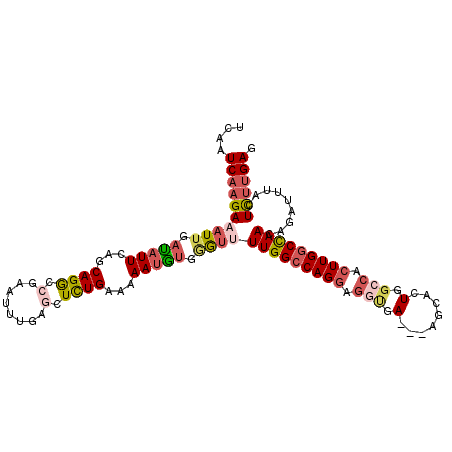
>3L_DroMel_CAF1 1996826 110 - 23771897 UCAAUCAUGAAAUUGACAUUCACCAGACCGAAUUCGAGCUCUGAAAAAUGUGGAUUUUGGCCAGGAGGUGAAGAAGCACUGGCCACUUGGCCAACAGAUUUACUCUUGAG ..((((..((((((.(((((...(((((((....)).).))))...))))).))))))(((((((.(((.(........).))).)))))))....)))).......... ( -32.10) >DroSec_CAF1 47689 107 - 1 UCAAUCAAGAUAUUGAUAUUCAGCAGGCCGAAUUUGAGCUCUGAAAAAUGUCGGUUUUGGCCAGGAGGUGA---AGCACUGGCCACUUGGCCAAAAGAUUUACUCUUGAG ....((((((..((((((((...(((((((....)).)).)))...))))))))(((((((((((.(((.(---.....).))).))))))))))).......)))))). ( -37.10) >DroSim_CAF1 62520 107 - 1 UCAAUCAAGAAAUUGAUAUUCAGCAGGCCUAAUUCGAGGUCUGAAAAAUGUGGGUUUUGGCCAGGAGGUGA---AGCACUGUCCACUUGGCCAACAGAUUUACUCUUGAG ....((((((((((.(((((...(((((((......)))))))...))))).))))(((((((((.((...---........)).))))))))).........)))))). ( -36.80) >DroEre_CAF1 53678 107 - 1 UCAAUCAAGAAAUUGAUAUUCAGCAGGCAGAAUUUGAGCUUUGAAAAAUACGAAAAUUGGACAGAAAAUUA---AAUACUGGCCACUUGGCCAACACAUUUACUUUCGAG ...(((((....))))).(((((..(((.........)))))))).....((((((((........)))).---.....(((((....)))))...........)))).. ( -19.00) >DroYak_CAF1 48800 107 - 1 UCAAUCAAGAAAUUUAUAUUCAGCAGGCAGAAUUUGGGCUUUGAAAAACAUGUGUUUUGGCCAGGAGACGA---AACACUGGCCUCUUGGCUAACACAUUUACUCUUGAG ....((((((........(((((.((.(........).)))))))....((((((..((((((((((.((.---.....))..))))))))))))))))....)))))). ( -31.20) >consensus UCAAUCAAGAAAUUGAUAUUCAGCAGGCCGAAUUUGAGCUCUGAAAAAUGUGGGUUUUGGCCAGGAGGUGA___AGCACUGGCCACUUGGCCAACAGAUUUACUCUUGAG ....((((((((((.(((((...((((.(........).))))...))))).))))(((((((((.(((.(........).))).))))))))).........)))))). (-17.84 = -19.44 + 1.60)

| Location | 1,996,863 – 1,996,973 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 85.39 |
| Mean single sequence MFE | -26.68 |
| Consensus MFE | -19.29 |
| Energy contribution | -20.57 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1996863 110 + 23771897 CUUCACCUCCUGGCCAAAAUCCACAUUUUUCAGAGCUCGAAUUCGGUCUGGUGAAUGUCAAUUUCAUGAUUGAAUGUGACACAUUCUUUAUGGGCCGGGAAUAUUAUAGA .......((((((((.((((..((((((.(((((.(.((....)))))))).))))))..))))(((((..(((((.....))))).))))))))))))).......... ( -35.70) >DroSec_CAF1 47725 108 + 1 --UCACCUCCUGGCCAAAACCGACAUUUUUCAGAGCUCAAAUUCGGCCUGCUGAAUAUCAAUAUCUUGAUUGAAUGUGGCACAUUCUUUAUGGGCCGGGACUAUUAUAGA --.....((((((((....((.((((((.(((((......((((((....)))))).......)).)))..))))))))..(((.....))))))))))).......... ( -30.32) >DroSim_CAF1 62556 108 + 1 --UCACCUCCUGGCCAAAACCCACAUUUUUCAGACCUCGAAUUAGGCCUGCUGAAUAUCAAUUUCUUGAUUGAAUGUGGCACAUUCUUUAUGGGCCGGGAAUAUUAUAGA --.....((((((((.....((((((..(((((.(((......)))....)))))..((((((....))))))))))))..(((.....))))))))))).......... ( -32.90) >DroEre_CAF1 53714 108 + 1 --UAAUUUUCUGUCCAAUUUUCGUAUUUUUCAAAGCUCAAAUUCUGCCUGCUGAAUAUCAAUUUCUUGAUUGAAUGUGGUACAUUCUUUAUGGGCCUGUAAUAUUGUAGA --.........(((((......((((((..((..((.........)).))..)))))).............(((((.....)))))....)))))............... ( -13.00) >DroYak_CAF1 48836 108 + 1 --UCGUCUCCUGGCCAAAACACAUGUUUUUCAAAGCCCAAAUUCUGCCUGCUGAAUAUAAAUUUCUUGAUUGAAUGUGGCACAUUCUUUAUGGGCCUGGAAUAUUGUAAA --.....(((.(((((((((....))))).....(((((.((((........))))......(((......))))).)))............)))).))).......... ( -21.50) >consensus __UCACCUCCUGGCCAAAACCCACAUUUUUCAGAGCUCAAAUUCGGCCUGCUGAAUAUCAAUUUCUUGAUUGAAUGUGGCACAUUCUUUAUGGGCCGGGAAUAUUAUAGA .......((((((((......(((((((.((((.......((((((....)))))).........))))..)))))))...(((.....))))))))))).......... (-19.29 = -20.57 + 1.28)

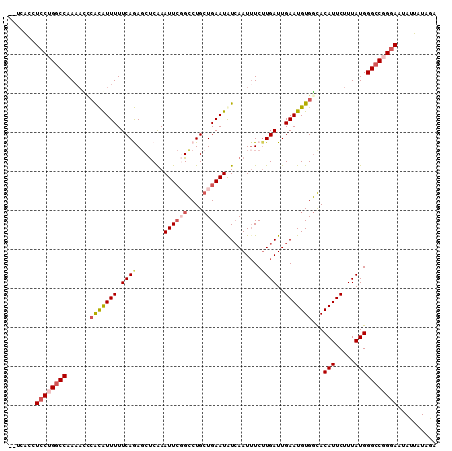
| Location | 1,996,863 – 1,996,973 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 85.39 |
| Mean single sequence MFE | -26.54 |
| Consensus MFE | -18.56 |
| Energy contribution | -19.48 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
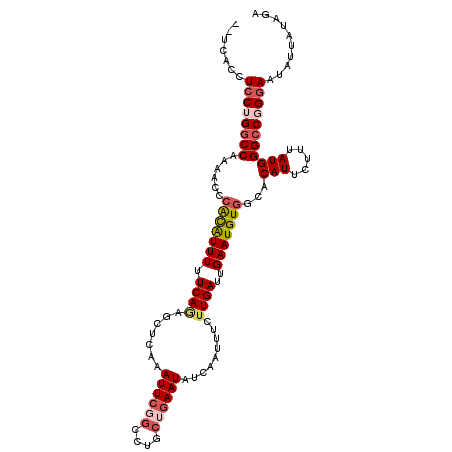
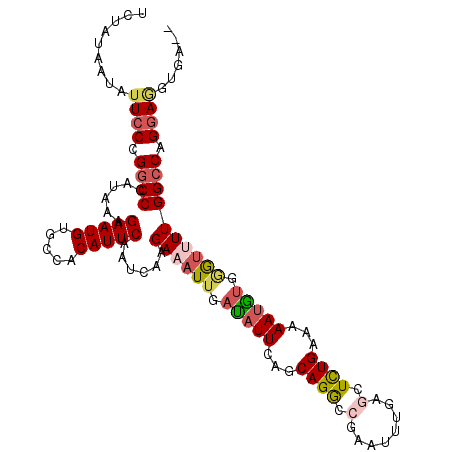
>3L_DroMel_CAF1 1996863 110 - 23771897 UCUAUAAUAUUCCCGGCCCAUAAAGAAUGUGUCACAUUCAAUCAUGAAAUUGACAUUCACCAGACCGAAUUCGAGCUCUGAAAAAUGUGGAUUUUGGCCAGGAGGUGAAG .........((((.((((......(((((.....)))))......((((((.(((((...(((((((....)).).))))...))))).)))))))))).))))...... ( -30.70) >DroSec_CAF1 47725 108 - 1 UCUAUAAUAGUCCCGGCCCAUAAAGAAUGUGCCACAUUCAAUCAAGAUAUUGAUAUUCAGCAGGCCGAAUUUGAGCUCUGAAAAAUGUCGGUUUUGGCCAGGAGGUGA-- ..........(((.((((......(((((.....)))))......((.(((((((((...(((((((....)).)).)))...))))))))).)))))).))).....-- ( -28.20) >DroSim_CAF1 62556 108 - 1 UCUAUAAUAUUCCCGGCCCAUAAAGAAUGUGCCACAUUCAAUCAAGAAAUUGAUAUUCAGCAGGCCUAAUUCGAGGUCUGAAAAAUGUGGGUUUUGGCCAGGAGGUGA-- .........((((.((((((((.....))))(((((((..(((((....)))))......(((((((......)))))))...))))))).....)))).))))....-- ( -35.70) >DroEre_CAF1 53714 108 - 1 UCUACAAUAUUACAGGCCCAUAAAGAAUGUACCACAUUCAAUCAAGAAAUUGAUAUUCAGCAGGCAGAAUUUGAGCUUUGAAAAAUACGAAAAUUGGACAGAAAAUUA-- (((............(((......(((((.....))))).(((((....)))))........)))....((..(..((((.......))))..)..)).)))......-- ( -16.60) >DroYak_CAF1 48836 108 - 1 UUUACAAUAUUCCAGGCCCAUAAAGAAUGUGCCACAUUCAAUCAAGAAAUUUAUAUUCAGCAGGCAGAAUUUGGGCUUUGAAAAACAUGUGUUUUGGCCAGGAGACGA-- .........((((.((((((((..(((((.....)))))................(((((.((.(........).))))))).....))))....)))).))))....-- ( -21.50) >consensus UCUAUAAUAUUCCCGGCCCAUAAAGAAUGUGCCACAUUCAAUCAAGAAAUUGAUAUUCAGCAGGCCGAAUUUGAGCUCUGAAAAAUGUGGGUUUUGGCCAGGAGGUGA__ .........((((.((((......(((((.....)))))......((((((.(((((...((((.(........).))))...))))).)))))))))).))))...... (-18.56 = -19.48 + 0.92)

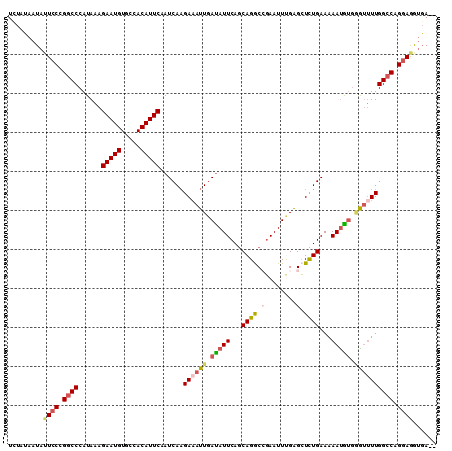
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:11 2006