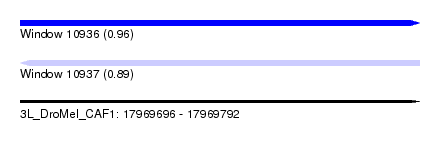
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,969,696 – 17,969,792 |
| Length | 96 |
| Max. P | 0.955496 |

| Location | 17,969,696 – 17,969,792 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 77.93 |
| Mean single sequence MFE | -24.45 |
| Consensus MFE | -14.07 |
| Energy contribution | -14.52 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
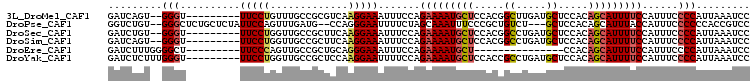
>3L_DroMel_CAF1 17969696 96 + 23771897 GAUCAGU--GGGU---------UUCCUGUUUGCCGCGUCAAGGAAAUUUCCAGAAAAUGCUCCACGGCUUGAUGCUCCACAGCAUUUUCCAUUUCCCCAUUAAAUCC ....(((--((((---------(((((.............))))))......(((((((((....(((.....)))....)))))))))......))))))...... ( -25.02) >DroPse_CAF1 99469 100 + 1 GGUCUGU--GGGCUCUGCUCUAUUCCAGUUUGAUG--CCAGGGAAUUUUCUAGCAAAUUUCCCGCUGUCU---GCUCCACAGCAUUUACCAUUUCCCCCCACCGUCC ((.(.((--(((...((((..(((((.((.....)--)...))))).....))))........(((((..---.....)))))..............))))).).)) ( -24.00) >DroSec_CAF1 93335 96 + 1 GAUCUGU--GGGU---------UUCCUGGUUGCCGCUUCAAGGAAAUUUCCAGAAAAUGCUCCACGGCCUGAUGCUCCACAGCAUUUUCCAUUUCCCCAUUAAAUCC (((..((--((((---------(((((((........)).))))))......(((((((((....(((.....)))....)))))))))......)))))...))). ( -24.70) >DroSim_CAF1 100639 96 + 1 GAUCAGU--GGGU---------UUCCUGGUUGCCGCUUCAAGGAAAUUUCCAGAAAAUGCUCCACGGCCUGAUGCUCCACAGCAUUUUCCAUUUCCCCAUUAAAUCC ....(((--((((---------(((((((........)).))))))......(((((((((....(((.....)))....)))))))))......))))))...... ( -24.50) >DroEre_CAF1 94645 83 + 1 GAUCUUUGGGGCU---------UUCCCAGUUGCCGCUGCAGGGAAAUUUCCAGAAAAUGCU---------------CCACAGCAUUUUCCAUUUCCCCAUUAAAUCC (((...(((((.(---------(((((...(((....)))))))))......(((((((((---------------....))))))))).....)))))....))). ( -26.10) >DroYak_CAF1 98811 98 + 1 GAUCUCUUUGGGU---------UUCCUGGUUGCCGCUCCAAGGAAUUUUCCAGAAAAUGCUCCACCGCCUGAUGCUCCACAGCAUUUUCCAUUUCCCCAUUAAAUCC (((.....((((.---------(((((((........)).))))).......(((((((((.....((.....)).....)))))))))......))))....))). ( -22.40) >consensus GAUCUGU__GGGU_________UUCCUGGUUGCCGCUUCAAGGAAAUUUCCAGAAAAUGCUCCACGGCCUGAUGCUCCACAGCAUUUUCCAUUUCCCCAUUAAAUCC .........(((..........(((((.............))))).......(((((((((.....((.....)).....)))))))))......)))......... (-14.07 = -14.52 + 0.45)
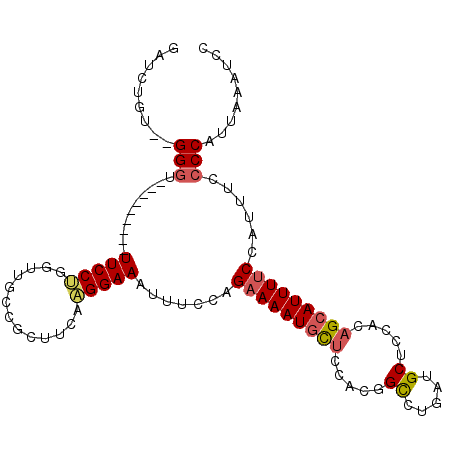
| Location | 17,969,696 – 17,969,792 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.93 |
| Mean single sequence MFE | -28.05 |
| Consensus MFE | -17.78 |
| Energy contribution | -18.28 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17969696 96 - 23771897 GGAUUUAAUGGGGAAAUGGAAAAUGCUGUGGAGCAUCAAGCCGUGGAGCAUUUUCUGGAAAUUUCCUUGACGCGGCAAACAGGAA---------ACCC--ACUGAUC .((((...((((.....((((((((((.((..((.....))..)).)))))))))).....((((((((......))...)))))---------))))--)..)))) ( -25.90) >DroPse_CAF1 99469 100 - 1 GGACGGUGGGGGGAAAUGGUAAAUGCUGUGGAGC---AGACAGCGGGAAAUUUGCUAGAAAAUUCCCUGG--CAUCAAACUGGAAUAGAGCAGAGCCC--ACAGACC ((...((((((((((.(((((((((((((.....---..))))).....)))))))).....)))))...--.......(((........)))..)))--))...)) ( -28.20) >DroSec_CAF1 93335 96 - 1 GGAUUUAAUGGGGAAAUGGAAAAUGCUGUGGAGCAUCAGGCCGUGGAGCAUUUUCUGGAAAUUUCCUUGAAGCGGCAACCAGGAA---------ACCC--ACAGAUC .(((((..((((.....((((((((((.((..((.....))..)).)))))))))).....((((((....(.(....)))))))---------))))--).))))) ( -26.50) >DroSim_CAF1 100639 96 - 1 GGAUUUAAUGGGGAAAUGGAAAAUGCUGUGGAGCAUCAGGCCGUGGAGCAUUUUCUGGAAAUUUCCUUGAAGCGGCAACCAGGAA---------ACCC--ACUGAUC .((((...((((.....((((((((((.((..((.....))..)).)))))))))).....((((((....(.(....)))))))---------))))--)..)))) ( -26.70) >DroEre_CAF1 94645 83 - 1 GGAUUUAAUGGGGAAAUGGAAAAUGCUGUGG---------------AGCAUUUUCUGGAAAUUUCCCUGCAGCGGCAACUGGGAA---------AGCCCCAAAGAUC .(((((..(((((....((((((((((....---------------)))))))))).....(((((((((....)))...)))))---------).))))).))))) ( -30.60) >DroYak_CAF1 98811 98 - 1 GGAUUUAAUGGGGAAAUGGAAAAUGCUGUGGAGCAUCAGGCGGUGGAGCAUUUUCUGGAAAAUUCCUUGGAGCGGCAACCAGGAA---------ACCCAAAGAGAUC .(((((..((((.....((((((((((.((..((.....))..)).))))))))))......(((((.((........)))))))---------.))))...))))) ( -30.40) >consensus GGAUUUAAUGGGGAAAUGGAAAAUGCUGUGGAGCAUCAGGCCGUGGAGCAUUUUCUGGAAAUUUCCUUGAAGCGGCAACCAGGAA_________ACCC__ACAGAUC .........((((((((((((((((((.((..((.....))..)).))))))))))....))))))))....................................... (-17.78 = -18.28 + 0.50)

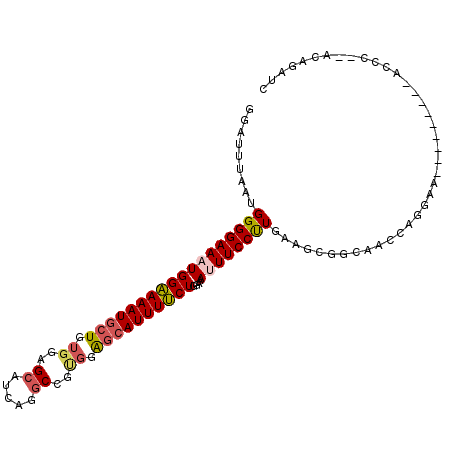
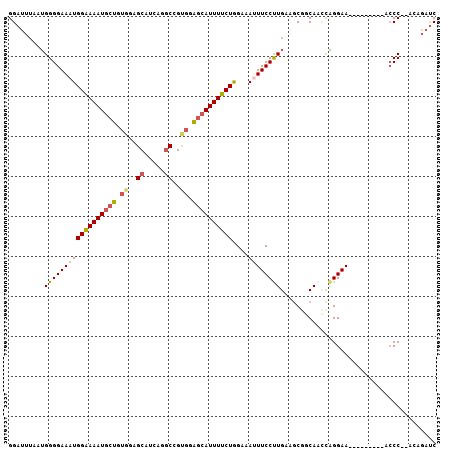
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:29 2006