
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,968,189 – 17,968,326 |
| Length | 137 |
| Max. P | 0.999993 |

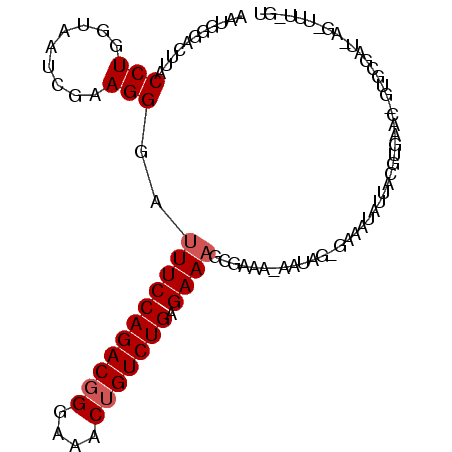
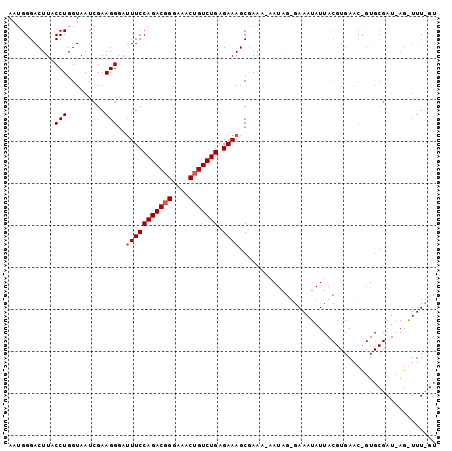
| Location | 17,968,189 – 17,968,291 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 65.36 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -15.80 |
| Energy contribution | -16.47 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.62 |
| SVM decision value | 5.77 |
| SVM RNA-class probability | 0.999993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17968189 102 - 23771897 AUUGGGACUUACCUGGUAAUCGAAGGCAUUUCCAGACGGGAAACUGUCUGAGAAAGCGAAAGAAUAGUGAAAUAUUACGUGAACUGUGCGAUUAGAUUUUGU .(..((.....))..)((((((.((((.(((((((((((....))))))).)))))).........((((....)))).....))...))))))........ ( -29.70) >DroSim_CAF1 99215 102 - 1 AAUGGGACUUACCUGGUAAUAGAAGGGAUUUCCAGACUGGAAACUGUCUGAGAAAGCGAAANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN ...........(((.........)))..(((((((((.(....).))))).))))............................................... ( -14.70) >DroEre_CAF1 93275 101 - 1 AAGGGGACUUACCUGGUAAUCGAAGGGG-UUCCAGACGGGAAACUGUCUGGGAAAGCAAAACAAUAGGGAAAUAUUACGUGAACCGUGCGAUAAGGUUUCGU (((....))).((((....((....)).-((((((((((....))))))))))...........))))(((((.((((((.......))).))).))))).. ( -32.40) >consensus AAUGGGACUUACCUGGUAAUCGAAGGGAUUUCCAGACGGGAAACUGUCUGAGAAAGCGAAA_AAUAG_GAAAUAUUACGUGAAC_GUGCGAU_AG_UUU_GU ...........(((.........)))..(((((((((((....))))))).))))............................................... (-15.80 = -16.47 + 0.67)

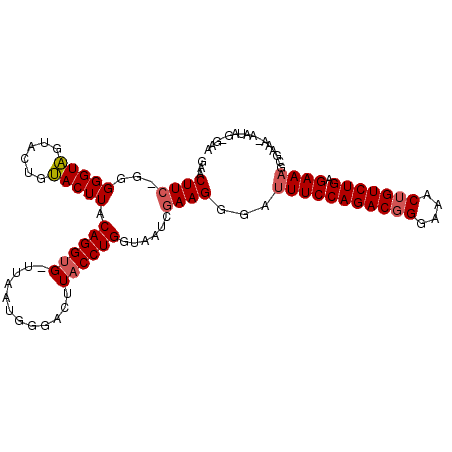
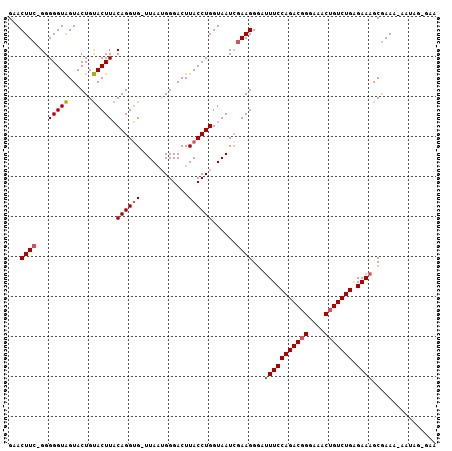
| Location | 17,968,220 – 17,968,326 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 82.81 |
| Mean single sequence MFE | -33.60 |
| Consensus MFE | -23.18 |
| Energy contribution | -24.07 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.984063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17968220 106 - 23771897 GAACUUCAGGGGGUAGUACUGUACUUACAGGUG-UUAUUGGGACUUACCUGGUAAUCGAAGGCAUUUCCAGACGGGAAACUGUCUGAGAAAGCGAAAGAAUAGUGAA ...((((.(..((((......))))..)....(-((((..((.....))..))))).))))((.(((((((((((....))))))).)))))).............. ( -34.10) >DroSim_CAF1 99246 106 - 1 GAACUUCUGGGGGUAGUACUGUACUUACAGGUG-UUAAUGGGACUUACCUGGUAAUAGAAGGGAUUUCCAGACUGGAAACUGUCUGAGAAAGCGAAANNNNNNNNNN ...((((((.(((((......))))).((((((-...........))))))....))))))...(((((((((.(....).))))).))))................ ( -27.30) >DroEre_CAF1 93306 105 - 1 GAACUUG-GGGGGUGGUACUGCACUUCCAGGGGAUUAAGGGGACUUACCUGGUAAUCGAAGGGG-UUCCAGACGGGAAACUGUCUGGGAAAGCAAAACAAUAGGGAA ...((..-(((((((......)))))))..))...((((....))))((((....((....)).-((((((((((....))))))))))...........))))... ( -39.40) >consensus GAACUUC_GGGGGUAGUACUGUACUUACAGGUG_UUAAUGGGACUUACCUGGUAAUCGAAGGGAUUUCCAGACGGGAAACUGUCUGAGAAAGCGAAA_AAUAG_GAA ...((((...(((((......))))).((((((............))))))......))))...(((((((((((....))))))).))))................ (-23.18 = -24.07 + 0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:27 2006