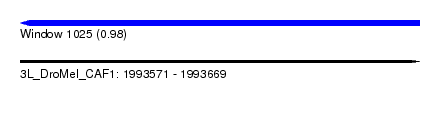
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,993,571 – 1,993,669 |
| Length | 98 |
| Max. P | 0.977294 |

| Location | 1,993,571 – 1,993,669 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.19 |
| Mean single sequence MFE | -28.19 |
| Consensus MFE | -18.88 |
| Energy contribution | -19.60 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
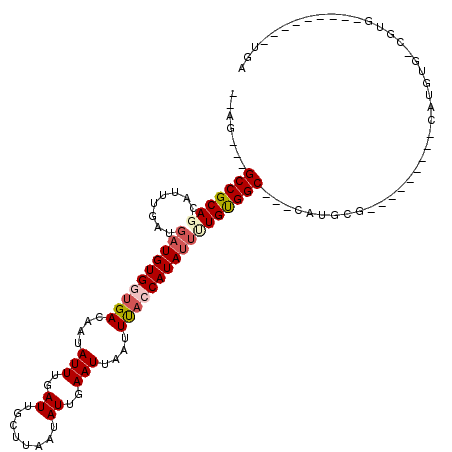
>3L_DroMel_CAF1 1993571 98 - 23771897 --AG---GCCGCAGCAUUUGAUGAUGUGGCGACAAUAUUUGAUUGCUUAAUAUUGAAUUAAUUUACCAUAUUUUGCGGC---CAUGCGGC-----GACAUGUG-CGUG--------CUGA --.(---(((((((........(((((((..(....(((..((........))..)))....)..))))))))))))))---)...((((-----.((.....-.)))--------))). ( -28.80) >DroPse_CAF1 58316 112 - 1 --AGGCAGCCGCAGAAUUUGAUGAUGUGGUGACAAUAUUUGAUUGCUUAAUAUUGAAUUAAUUUACCAUACGCUGUGGCAGACAUGCGGU-----G-CAUGUGGCAUGGUAGCAUAUUGU --..((.(((((((.......((.((((((((....(((..((........))..)))....))))))))))))))))).....(((.((-----(-(.....)))).)))))....... ( -32.11) >DroGri_CAF1 43096 90 - 1 GCCU---GCCGCAGCAGUUGAUGAUGUGGUGACAAUAUUUGAUUGCUUAAUAUUGAAUUAAUUUAGCAUAUU-UGUGGCGGCAAUGCG-------------UG-UGUG------------ ((.(---(((((((((((..(.....((....))....)..))))))..................(((....-))).))))))..)).-------------..-....------------ ( -26.20) >DroWil_CAF1 65500 90 - 1 --AA---GCCGCAGCAUUUGAUGAUGUGGUGACAAUAUUUGAUUGCUUAAUAUUGAAUUAAUUUACCAUAUUUUGUGGC---C-------------GCAUGUG-GGCG--------UUGA --..---(((((((........((((((((((....(((..((........))..)))....)))))))))))))))))---(-------------((.....-.)))--------.... ( -24.20) >DroMoj_CAF1 51463 88 - 1 --CU---GCCGCAGCUGUUGAUGAUGUGGUGACAAUAUUUGAUUGCGUAAUAUUGAAUUAAUUCAACAUAUU-UGUGGCCGCAAUGCG-------------UG-UGCG------------ --..---(((((((.((((((.....((((..(((((((((....)).))))))).))))..))))))...)-))))))((((.....-------------..-))))------------ ( -22.10) >DroAna_CAF1 41051 103 - 1 --AG---GCCGCAGCAUUUGAUGAUGUGGUGACAAUAUUUGAUUGCUUAAUAUUGAAUUAAUUUACCAUAUUUUGCGGC---CAUGUGGCGGUGCGGCUUGUG-CGUG--------CUGA --.(---(((((((........((((((((((....(((..((........))..)))....)))))))))))))))))---)......(((..((.(....)-))..--------))). ( -35.70) >consensus __AG___GCCGCAGCAUUUGAUGAUGUGGUGACAAUAUUUGAUUGCUUAAUAUUGAAUUAAUUUACCAUAUUUUGUGGC___CAUGCG_________CAUGUG_CGUG_________UGA .......(((((((........((((((((((....(((..((........))..)))....)))))))))))))))))......................................... (-18.88 = -19.60 + 0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:08 2006