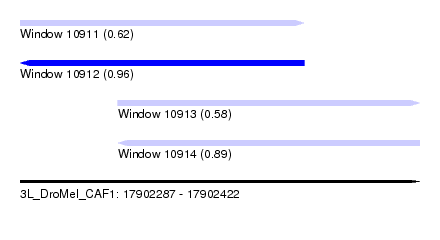
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
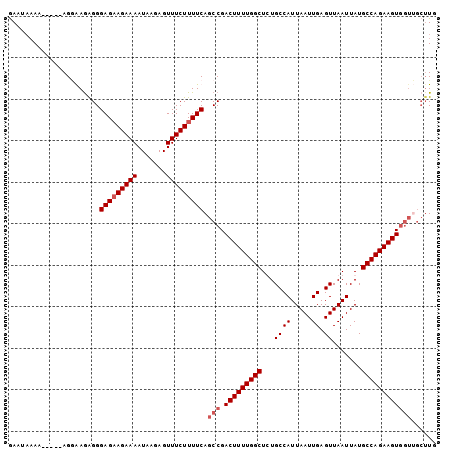
| Location | 17,902,287 – 17,902,422 |
| Length | 135 |
| Max. P | 0.960705 |

| Location | 17,902,287 – 17,902,383 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 87.23 |
| Mean single sequence MFE | -14.45 |
| Consensus MFE | -11.02 |
| Energy contribution | -11.28 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17902287 96 + 23771897 CA-------ACUUCUGGCAUAAUUAACUCAAUUAAUGGCAGAGCCAAAAGUCGGCUGAAAAGAAACUCUUAUUUUCUACUCCCUCUCUCUUCUUUUUUUUUUU ..-------...((((.(((((((.....)))).))).))))(((.......))).((((((((.........................))))))))...... ( -13.41) >DroSec_CAF1 26020 94 + 1 CAAGCAACCACUUCUGGCAUAAUUAACUCAAUUAAUGGCAAAGCCAAAAGUCGGCUGAAAAGAAACUCUUAUUUUCUUCUCCCUCUUCCU-----UU----UC ..(((....((((.((((...(((((.....)))))......)))).))))..)))((.((((((.......)))))).)).........-----..----.. ( -13.90) >DroSim_CAF1 26289 98 + 1 CAAGCAACCACUUCUGGCAUAAUUAACUCAAUUAAUGGCAAAGCCAAAAGUCGGCUGAAAAGAAACUCUUAUUUUCUUCUCCCUUUUCCU-----UUUUAUUC ..(((....((((.((((...(((((.....)))))......)))).))))..)))((.((((((.......)))))).)).........-----........ ( -13.90) >DroEre_CAF1 27259 98 + 1 CAAGCCACCACUUCUGGCAUAAUUAACUCAAUUAAUGGCAGAGCCAAAAGUCGGCUGAAAAGAAACUCUUAUUUUCUUCUCCCCCUCGCU-----UUUUAUUC .((((.......((((.(((((((.....)))).))).))))(((.......))).((.((((((.......)))))).))......)))-----)....... ( -16.60) >consensus CAAGCAACCACUUCUGGCAUAAUUAACUCAAUUAAUGGCAAAGCCAAAAGUCGGCUGAAAAGAAACUCUUAUUUUCUUCUCCCUCUCCCU_____UUUUAUUC .............(((((.(((((.....))))).((((...))))...)))))..((.((((((.......)))))).))...................... (-11.02 = -11.28 + 0.25)

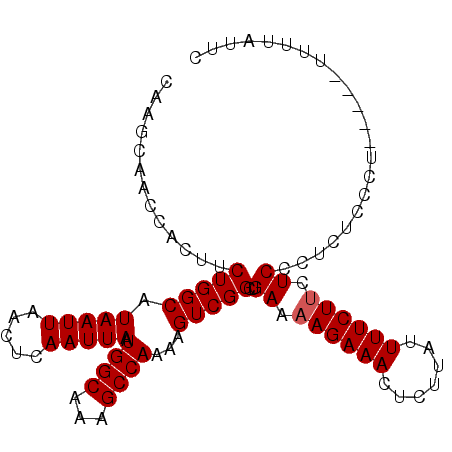
| Location | 17,902,287 – 17,902,383 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 87.23 |
| Mean single sequence MFE | -24.98 |
| Consensus MFE | -19.83 |
| Energy contribution | -20.82 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17902287 96 - 23771897 AAAAAAAAAAAGAAGAGAGAGGGAGUAGAAAAUAAGAGUUUCUUUUCAGCCGACUUUUGGCUCUGCCAUUAAUUGAGUUAAUUAUGCCAGAAGU-------UG ...........(((((((((.....((.....))....)))))))))...(((((((((((...((((.....)).)).......)))))))))-------)) ( -21.80) >DroSec_CAF1 26020 94 - 1 GA----AA-----AGGAAGAGGGAGAAGAAAAUAAGAGUUUCUUUUCAGCCGACUUUUGGCUUUGCCAUUAAUUGAGUUAAUUAUGCCAGAAGUGGUUGCUUG ..----..-----.....(((.(((((((((.......)))))))))((((.(((((((((...((((.....)).)).......))))))))))))).))). ( -25.30) >DroSim_CAF1 26289 98 - 1 GAAUAAAA-----AGGAAAAGGGAGAAGAAAAUAAGAGUUUCUUUUCAGCCGACUUUUGGCUUUGCCAUUAAUUGAGUUAAUUAUGCCAGAAGUGGUUGCUUG ........-----.....(((.(((((((((.......)))))))))((((.(((((((((...((((.....)).)).......))))))))))))).))). ( -24.90) >DroEre_CAF1 27259 98 - 1 GAAUAAAA-----AGCGAGGGGGAGAAGAAAAUAAGAGUUUCUUUUCAGCCGACUUUUGGCUCUGCCAUUAAUUGAGUUAAUUAUGCCAGAAGUGGUGGCUUG .......(-----(((......(((((((((.......))))))))).(((.(((((((((...((((.....)).)).......)))))))))))).)))). ( -27.90) >consensus GAAUAAAA_____AGGAAGAGGGAGAAGAAAAUAAGAGUUUCUUUUCAGCCGACUUUUGGCUCUGCCAUUAAUUGAGUUAAUUAUGCCAGAAGUGGUUGCUUG ......................(((((((((.......))))))))).(((.(((((((((...((((.....)).)).......))))))))))))...... (-19.83 = -20.82 + 1.00)

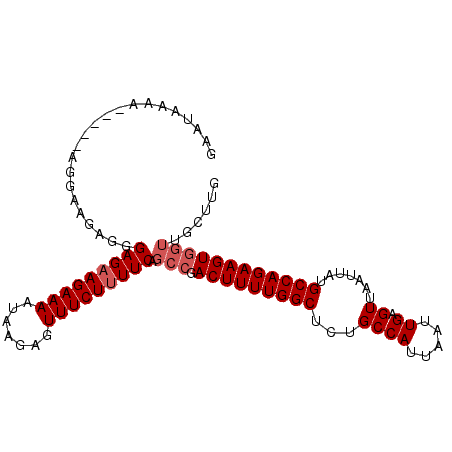
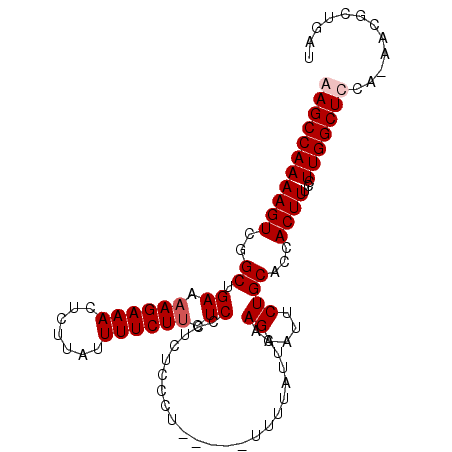
| Location | 17,902,320 – 17,902,422 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 89.37 |
| Mean single sequence MFE | -16.05 |
| Consensus MFE | -13.38 |
| Energy contribution | -14.12 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582116 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17902320 102 + 23771897 GAGCCAAAAGUCGGCUGAAAAGAAACUCUUAUUUUCUACUCCCUCUCUCUUCUUUUUUUUUUUAAGAAUUCUGCAACACUUUCCUUGGCUCCA-AACGCUGAU (((((((((((..((.(((.(((((.......)))))............(((((.........)))))))).))...))))...)))))))..-......... ( -18.40) >DroSec_CAF1 26060 94 + 1 AAGCCAAAAGUCGGCUGAAAAGAAACUCUUAUUUUCUUCUCCCUCUUCCU-----UU----UCAAGAAUUCUGCACCACUUUCCUUGGCUCCAAAACGCUGAU .((((((((((.(((.(((((((((.......)))))).....((((...-----..----..)))).))).)).).))))...))))))............. ( -14.80) >DroSim_CAF1 26329 98 + 1 AAGCCAAAAGUCGGCUGAAAAGAAACUCUUAUUUUCUUCUCCCUUUUCCU-----UUUUAUUCAAGAAUUCUGCACCACUUUCCUUGGCUCCAAAACGCUGAU .((((((((((.(((.(((((((((.......))))))..........((-----(.......)))..))).)).).))))...))))))............. ( -13.40) >DroEre_CAF1 27299 97 + 1 GAGCCAAAAGUCGGCUGAAAAGAAACUCUUAUUUUCUUCUCCCCCUCGCU-----UUUUAUUCAAGAAUUCUGCACCACUUUCCUUGGCUCCA-AGCGCUGAU (((((((((((.((..((.((((((.......)))))).))..))..((.-----((((.....))))....))...))))...)))))))..-......... ( -17.60) >consensus AAGCCAAAAGUCGGCUGAAAAGAAACUCUUAUUUUCUUCUCCCUCUCCCU_____UUUUAUUCAAGAAUUCUGCACCACUUUCCUUGGCUCCA_AACGCUGAU (((((((((((..((.((.((((((.......)))))).)).......................((....))))...))))...)))))))............ (-13.38 = -14.12 + 0.75)

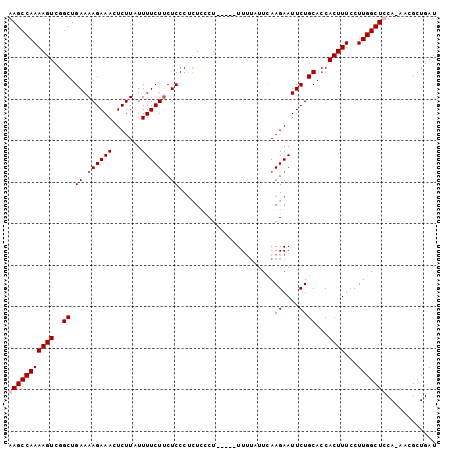
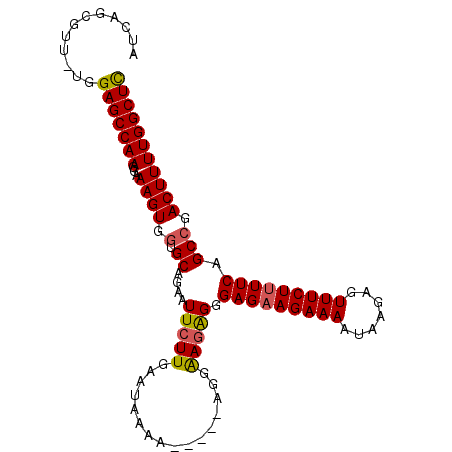
| Location | 17,902,320 – 17,902,422 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 89.37 |
| Mean single sequence MFE | -24.50 |
| Consensus MFE | -20.48 |
| Energy contribution | -20.54 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17902320 102 - 23771897 AUCAGCGUU-UGGAGCCAAGGAAAGUGUUGCAGAAUUCUUAAAAAAAAAAAGAAGAGAGAGGGAGUAGAAAAUAAGAGUUUCUUUUCAGCCGACUUUUGGCUC .........-..(((((((...((((...((....(((((.........)))))..(((((((((.............))))))))).))..))))))))))) ( -19.82) >DroSec_CAF1 26060 94 - 1 AUCAGCGUUUUGGAGCCAAGGAAAGUGGUGCAGAAUUCUUGA----AA-----AGGAAGAGGGAGAAGAAAAUAAGAGUUUCUUUUCAGCCGACUUUUGGCUU ............(((((((...((((.(.((....(((((..----..-----)))))....(((((((((.......))))))))).))).))))))))))) ( -26.70) >DroSim_CAF1 26329 98 - 1 AUCAGCGUUUUGGAGCCAAGGAAAGUGGUGCAGAAUUCUUGAAUAAAA-----AGGAAAAGGGAGAAGAAAAUAAGAGUUUCUUUUCAGCCGACUUUUGGCUU ............(((((((...((((.(.((....(((((........-----)))))....(((((((((.......))))))))).))).))))))))))) ( -25.20) >DroEre_CAF1 27299 97 - 1 AUCAGCGCU-UGGAGCCAAGGAAAGUGGUGCAGAAUUCUUGAAUAAAA-----AGCGAGGGGGAGAAGAAAAUAAGAGUUUCUUUUCAGCCGACUUUUGGCUC .........-..(((((((...((((.(.((....((((((.......-----..)))))).(((((((((.......))))))))).))).))))))))))) ( -26.30) >consensus AUCAGCGUU_UGGAGCCAAGGAAAGUGGUGCAGAAUUCUUGAAUAAAA_____AGGAAGAGGGAGAAGAAAAUAAGAGUUUCUUUUCAGCCGACUUUUGGCUC ............(((((((...((((.(.((....(((((................))))).(((((((((.......))))))))).))).))))))))))) (-20.48 = -20.54 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:07 2006