
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,902,087 – 17,902,230 |
| Length | 143 |
| Max. P | 0.730325 |

| Location | 17,902,087 – 17,902,206 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.98 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -18.72 |
| Energy contribution | -20.28 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
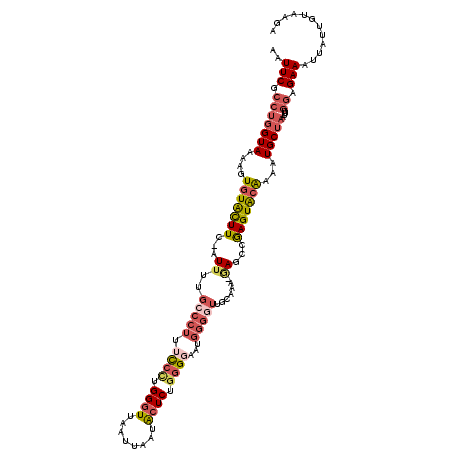
>3L_DroMel_CAF1 17902087 119 - 23771897 AAUUCGCCUGGUAAAAGUGUACUUC-AUUUUGCCCUUUCCUUGGGUUAAUUAAUACUCUGGGGAUUGGGGUUGAUUUGCAGCCGAGUACAAAAUGCUAAUUUGGAGAAAUUAUUGUAAGA ..(((.(((((((....(((((((.-.......((..((((..(((........)).)..))))..))(((((.....))))))))))))...)))))....)).)))............ ( -32.20) >DroSec_CAF1 25819 118 - 1 AAUUCGCCUGGUAAAAGUGUACUUC-AUUUUGCCCUUUCCCUGGGUUAAUUAAUACUCUGGGGAAUGGGCUUGCAAA-GAGCCGAGUACAAAAUGCUAAUUUGGAGAAAUUAUUGUAAGA ..(((.(((((((....(((((((.-.....((((.(((((..(((........)).)..))))).))))..((...-..)).)))))))...)))))....)).)))............ ( -34.50) >DroSim_CAF1 26088 118 - 1 AAUUCGCCUGGUAAAAGUGUACUUC-AUUUUGCCCUUUCCCUGGGUUAAUUAAUACUCUGGGGAAUGGGGUUGCAAA-GAGCCGAGUACAAAAUGCUAAUUUGGAGAAAUUAUUGUAAGA ..(((.(((((((....(((((((.-.((((((((((((((..(((........)).)..))))).)))))....))-))...)))))))...)))))....)).)))............ ( -32.60) >DroEre_CAF1 27055 117 - 1 AAUUCGCCUGGUAAAAGUGUGCUUC-AUUUUGCCCUUUCCCUGGGUUAAUUAGUACUCUGGGUACUGGGGUUGCAGA-GAGCCGAGUACAAAAUGCUAAUUUGGGGAAA-UAUUUUAAGA ..(((.(((((((....(((((((.-.(((((((((..(((.((((........)))).)))....)))))....))-))...)))))))...)))))....)).))).-.......... ( -31.70) >DroYak_CAF1 27467 117 - 1 AAUUCGCCUGGUAAAAGUGUACUUC-AUUUCGCCCUUUCCCUGGGUUAAUUAAUACUCAGGGUAAUGGGGUUGCAAA-GAGCCGAGUGCGAAAUGCUAAUUUGGGGAAA-UAUUUUAAGA ..(((.(((((((....(((((((.-.....(((((..((((((((........))))))))....))))).((...-..)).)))))))...)))))....)).))).-.......... ( -34.90) >DroAna_CAF1 24684 95 - 1 UAUUCGACGAGUAAAAGUCUAUUUCCAUUACUUUCAUUUCGGGGGUUAAUUAGAGGUCCAAGG---------------AAGUCAAGUAUGAAAUGCAAAUUUAAAGAAAA---------- ..((((((........)))((((((...(((((...((((..((..(.......)..))..))---------------))...))))).))))))..........)))..---------- ( -16.20) >consensus AAUUCGCCUGGUAAAAGUGUACUUC_AUUUUGCCCUUUCCCUGGGUUAAUUAAUACUCUGGGGAAUGGGGUUGCAAA_GAGCCGAGUACAAAAUGCUAAUUUGGAGAAAUUAUUGUAAGA ..(((.(((((((....(((((((...((..(((((.((((.((((........)))).))))...))))).......))...)))))))...)))))....)).)))............ (-18.72 = -20.28 + 1.56)

| Location | 17,902,127 – 17,902,230 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 79.22 |
| Mean single sequence MFE | -28.05 |
| Consensus MFE | -15.03 |
| Energy contribution | -18.20 |
| Covariance contribution | 3.17 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.54 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17902127 103 - 23771897 GGAGUAAGCAGACCAAGAAUUUGUAAUUCGCCUGGUAAAAGUGUACUUC-AUUUUGCCCUUUCCUUGGGUUAAUUAAUACUCUGGGGAUUGGGGUUGAUUUGCA ((((((.((..((((.((((.....))))...))))....)).))))))-..((.(((((.((((..(((........)).)..))))..))))).))...... ( -29.30) >DroSec_CAF1 25859 102 - 1 GGAGUAAGCAGACCAAGAAUUUGUAAUUCGCCUGGUAAAAGUGUACUUC-AUUUUGCCCUUUCCCUGGGUUAAUUAAUACUCUGGGGAAUGGGCUUGCAAA-GA ((((((.((..((((.((((.....))))...))))....)).))))))-.....((((.(((((..(((........)).)..))))).)))).......-.. ( -33.80) >DroSim_CAF1 26128 102 - 1 GCAGUAAGCAGACCAAGAAUUUGUAAUUCGCCUGGUAAAAGUGUACUUC-AUUUUGCCCUUUCCCUGGGUUAAUUAAUACUCUGGGGAAUGGGGUUGCAAA-GA ((((((.((..((((.((((.....))))...))))....)).)))...-.....((((((((((..(((........)).)..))))).))))))))...-.. ( -29.40) >DroEre_CAF1 27094 102 - 1 GGAGUAAGCAGACCAAGAAUUUGUAAUUCGCCUGGUAAAAGUGUGCUUC-AUUUUGCCCUUUCCCUGGGUUAAUUAGUACUCUGGGUACUGGGGUUGCAGA-GA ((((((.((..((((.((((.....))))...))))....)).))))))-.((((((((((.(((.((((........)))).)))....))))..)))))-). ( -29.00) >DroYak_CAF1 27506 102 - 1 GGAGUAAGCAGACCAAGAAUUUGUAAUUCGCCUGGUAAAAGUGUACUUC-AUUUCGCCCUUUCCCUGGGUUAAUUAAUACUCAGGGUAAUGGGGUUGCAAA-GA ((((((.((..((((.((((.....))))...))))....)).))))))-.....(((((..((((((((........))))))))....)))))......-.. ( -33.30) >DroAna_CAF1 24714 79 - 1 ----------AACCAAGAAUUCGUUAUUCGACGAGUAAAAGUCUAUUUCCAUUACUUUCAUUUCGGGGGUUAAUUAGAGGUCCAAGG---------------AA ----------.(((....(((((((....))))))).....((((........(((..(.....)..)))....)))))))......---------------.. ( -13.50) >consensus GGAGUAAGCAGACCAAGAAUUUGUAAUUCGCCUGGUAAAAGUGUACUUC_AUUUUGCCCUUUCCCUGGGUUAAUUAAUACUCUGGGGAAUGGGGUUGCAAA_GA ((((((.((..((((.((((.....))))...))))....)).))))))......(((((.((((.((((........)))).))))...)))))......... (-15.03 = -18.20 + 3.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:03 2006