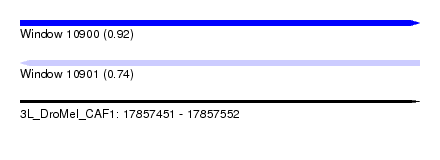
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,857,451 – 17,857,552 |
| Length | 101 |
| Max. P | 0.924381 |

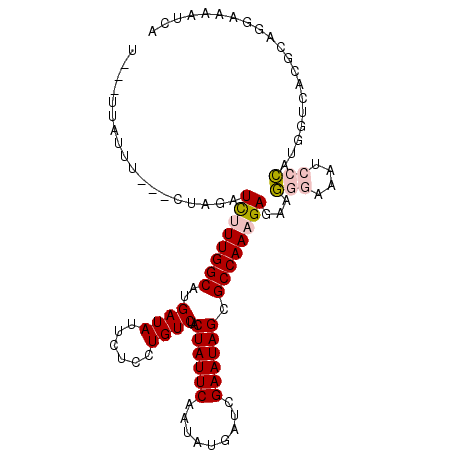
| Location | 17,857,451 – 17,857,552 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.29 |
| Mean single sequence MFE | -28.77 |
| Consensus MFE | -18.83 |
| Energy contribution | -19.50 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17857451 101 + 23771897 UGAUUUUCCUGCGUGACCAUGGGAUUCCCCUCCUCGUUGGCGCUAUUCGAUCAUAUUGAAUAGUAGACAGGAGAAUAUCAUGCCAAAGAUCUAA---AAAUAA---A .((((((...((((((....(((...)))(((((.(((...((((((((((...)))))))))).))))))))....))))))..))))))...---......---. ( -30.60) >DroPse_CAF1 20391 101 + 1 UGCUCUUGCAGCUG---CAACUCCUCGCCGAUCUCUUUGGCGCUAUUCAAUCAUAUUGAAUAGUAGACAAGAGAAUAUCAUGCCAAAAAUCUACGAAAAACGA---A ..((((((...(((---(.......((((((.....)))))).((((((((...)))))))))))).))))))..............................---. ( -24.00) >DroSec_CAF1 23258 101 + 1 UGAGUUUCCUGGGUGACCAUGGGAUUUCCCUCCUCGUUGGCGCUAUUCGAUCAUAUUGAAUAGUAGACAGGAGAAUAUCAUGCCAAAGAUCUAA---AAACAA---A .(((.(((((((....))).)))).))).(((((.(((...((((((((((...)))))))))).)))))))).....................---......---. ( -29.70) >DroEre_CAF1 24539 104 + 1 UGAUUUUCCUGCGUGACCAUGGGAUUUCCCUGCUCUUUGGCGCUAUUCGAUCAUAUUGAAUAGUAGACAGGAGAAUAUCAUGCCAAAGAUCUAG---CGAUAGGAAA ....((((((((((......(((....)))...((((((((((((((((((...))))))))))......((.....))..))))))))....)---)).))))))) ( -34.50) >DroYak_CAF1 40734 104 + 1 UGAUUUUCCUGCGUGACCAUGGGAUUUCCCUUCUCUUUGGCGCUAUUCGAUCAUAUUGAAUAGUAGACAGGAGAAUAUCAUGCCAAAGAUCUAG---AAAUAAGAAA .((((((...((((((....(((....)))(((((((.(..((((((((((...))))))))))...))))))))..))))))..))))))...---.......... ( -30.20) >DroPer_CAF1 20423 101 + 1 UGCUCUUGCAGCAG---CAACUCCUCGCCGAUCUCUUUGGCGCUAUUCAAUCAUAUUGAAUAGUAGACAAGAGAAUAUCAUGCCAAAAAUCUACGAAAAACGA---A (((....)))(((.---......((((((((.....)))))((((((((((...))))))))))......))).......)))....................---. ( -23.64) >consensus UGAUUUUCCUGCGUGACCAUGGGAUUUCCCUUCUCUUUGGCGCUAUUCGAUCAUAUUGAAUAGUAGACAGGAGAAUAUCAUGCCAAAGAUCUAA___AAACAA___A ....................(((....)))...((((((((((((((((((...))))))))))......((.....))..)))))))).................. (-18.83 = -19.50 + 0.67)

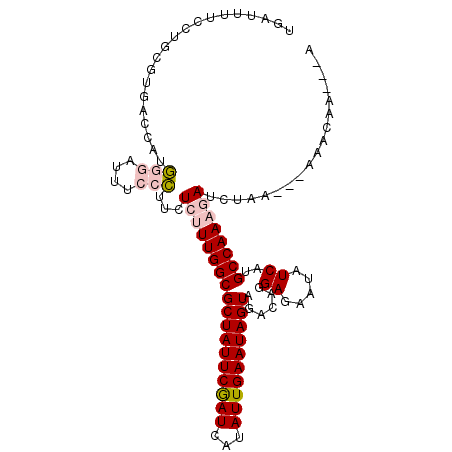
| Location | 17,857,451 – 17,857,552 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 81.29 |
| Mean single sequence MFE | -27.07 |
| Consensus MFE | -17.51 |
| Energy contribution | -17.85 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.735123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17857451 101 - 23771897 U---UUAUUU---UUAGAUCUUUGGCAUGAUAUUCUCCUGUCUACUAUUCAAUAUGAUCGAAUAGCGCCAACGAGGAGGGGAAUCCCAUGGUCACGCAGGAAAAUCA .---......---...(((.(((.((.((((...(((((((...((((((.........)))))).....)).)))))(((...)))...)))).))..))).))). ( -21.80) >DroPse_CAF1 20391 101 - 1 U---UCGUUUUUCGUAGAUUUUUGGCAUGAUAUUCUCUUGUCUACUAUUCAAUAUGAUUGAAUAGCGCCAAAGAGAUCGGCGAGGAGUUG---CAGCUGCAAGAGCA (---((.((((((((.((((((((((..((((......))))..(((((((((...))))))))).))))..)))))).))))))))(((---(....))))))).. ( -30.30) >DroSec_CAF1 23258 101 - 1 U---UUGUUU---UUAGAUCUUUGGCAUGAUAUUCUCCUGUCUACUAUUCAAUAUGAUCGAAUAGCGCCAACGAGGAGGGAAAUCCCAUGGUCACCCAGGAAACUCA .---..((((---((...((.(((((..((((......))))..((((((.........)))))).))))).))...(((....))).(((....)))))))))... ( -22.60) >DroEre_CAF1 24539 104 - 1 UUUCCUAUCG---CUAGAUCUUUGGCAUGAUAUUCUCCUGUCUACUAUUCAAUAUGAUCGAAUAGCGCCAAAGAGCAGGGAAAUCCCAUGGUCACGCAGGAAAAUCA ((((((...(---((((.((((((((..((((......))))..((((((.........)))))).)))))))).).(((....))).)))).....)))))).... ( -29.80) >DroYak_CAF1 40734 104 - 1 UUUCUUAUUU---CUAGAUCUUUGGCAUGAUAUUCUCCUGUCUACUAUUCAAUAUGAUCGAAUAGCGCCAAAGAGAAGGGAAAUCCCAUGGUCACGCAGGAAAAUCA ((((((....---.....((((((((..((((......))))..((((((.........)))))).))))))))...(((....)))..........)))))).... ( -25.90) >DroPer_CAF1 20423 101 - 1 U---UCGUUUUUCGUAGAUUUUUGGCAUGAUAUUCUCUUGUCUACUAUUCAAUAUGAUUGAAUAGCGCCAAAGAGAUCGGCGAGGAGUUG---CUGCUGCAAGAGCA .---..((((((.((((.((((((((..((((......))))..(((((((((...))))))))).))))))))...((((((....)))---))))))))))))). ( -32.00) >consensus U___UUAUUU___CUAGAUCUUUGGCAUGAUAUUCUCCUGUCUACUAUUCAAUAUGAUCGAAUAGCGCCAAAGAGAAGGGAAAUCCCAUGGUCACGCAGGAAAAUCA ..................((((((((..((((......))))..((((((.........)))))).))))))))...(((....))).................... (-17.51 = -17.85 + 0.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:54 2006