
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,853,809 – 17,853,941 |
| Length | 132 |
| Max. P | 0.901596 |

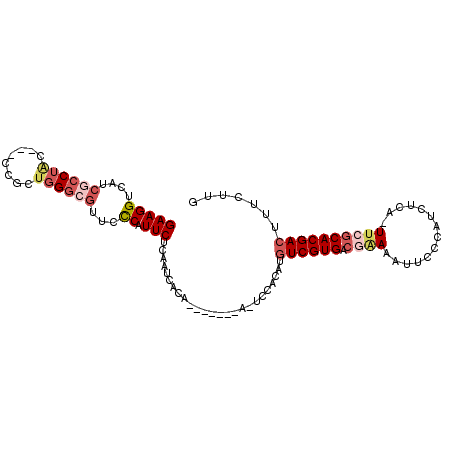
| Location | 17,853,809 – 17,853,910 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 77.63 |
| Mean single sequence MFE | -21.57 |
| Consensus MFE | -12.70 |
| Energy contribution | -14.09 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17853809 101 + 23771897 GAAGGUCACCUCCUAC---CCGCUGGGCGUUCCCAUUCUCAAUUACACAUGUACAAUCCACAUGUCGUGACGAAAAUACCCAUCUCA-UUCGCACGACUUUCUUG (((((((.........---....((((....))))...........((((((.......)))))).(((.((((.............-))))))))))))))... ( -21.72) >DroSec_CAF1 19437 94 + 1 GAAGGUCAUCGCCUAC---CCGCUGGGCGUUCUCAUUCCUAAUUACA-------AUUCCACAUGUCGUGACGAAAAUUCCCAUCUCA-UUCGCACGACUUUCUUG (((((((..((((((.---....))))))..................-------............(((.((((.............-))))))))))))))... ( -21.82) >DroSim_CAF1 23501 94 + 1 GAAGGUCAUCGCCUAC---CCGCUGGGCGUUCCCAUUCCCAAUUACA-------AUUCCACAUGUCGUGACGAAAAUUCCCAUCUCA-UUCGCACGACUUUCUUG (((((....((((((.---....))))))...)).))).........-------.........((((((.((((.............-))))))))))....... ( -21.82) >DroEre_CAF1 20961 93 + 1 GAAGGUCAUCGCCAAU---CCGCCGGGCUUUUCCAUUCUCAAUCGCA-------A-UCCACAUGUCGUGACAAAAACUCCCAUCUCA-UGCGCACGACUUUCCUG (((((((...(((...---......)))...................-------.-..(.((((..(((...........)))..))-)).)...)))))))... ( -15.70) >DroYak_CAF1 37300 94 + 1 GAAGGUCAUCGCCUAC---UCACUGGGCGUUCCCAUUCUCAAUCACA-------A-UCCACAUUUCGUGGGGAAAGCUUCCACGACUUUUCGCACGACUUUUUUG (((((((..((((((.---....))))))..................-------.-..........(((.(((((((......).)))))).))))))))))... ( -24.30) >DroAna_CAF1 17852 100 + 1 GAAGGUCAUCAGCUGGGCAACACUGGGCGGUUUCAUUCUCAGUCACA--GUCACAGUCCAAAAGUCGUGACUGAAAUUACCAUCCCA-UUCGCACGACACUUU-- (((((((........(....)..((((.(((...(((.((((((((.--.................))))))))))).)))..))))-.......))).))))-- ( -24.07) >consensus GAAGGUCAUCGCCUAC___CCGCUGGGCGUUCCCAUUCUCAAUCACA_______A_UCCACAUGUCGUGACGAAAAUUCCCAUCUCA_UUCGCACGACUUUCUUG (((((....((((((........))))))...)).))).........................((((((.((((..............))))))))))....... (-12.70 = -14.09 + 1.39)


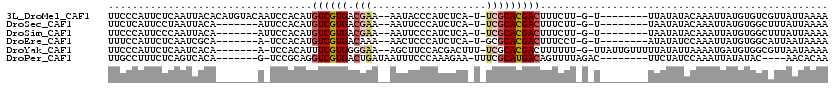
| Location | 17,853,835 – 17,853,941 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.66 |
| Mean single sequence MFE | -17.67 |
| Consensus MFE | -5.74 |
| Energy contribution | -6.33 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.32 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17853835 106 + 23771897 UUCCCAUUCUCAAUUACACAUGUACAAUCCACAUGUCGUGACGAA--AAUACCCAUCUCA-U-UCGCACGACUUUCUU-G-U--------UUAUAUACAAAUUAUGUGUCGUUAUUAAAA ...........(((.(((((((............((((((.((((--.............-)-)))))))))....((-(-(--------......))))..))))))).)))....... ( -19.82) >DroSec_CAF1 19463 99 + 1 UUCUCAUUCCUAAUUACA-------AUUCCACAUGUCGUGACGAA--AAUUCCCAUCUCA-U-UCGCACGACUUUCUU-G-U--------UAAUAUACAAAUUAUGUGGCUUUAUUAAAA .................(-------(..((((((((((((.((((--.............-)-)))))))))....((-(-(--------......))))...))))))..))....... ( -19.52) >DroSim_CAF1 23527 99 + 1 UUCCCAUUCCCAAUUACA-------AUUCCACAUGUCGUGACGAA--AAUUCCCAUCUCA-U-UCGCACGACUUUCUU-G-U--------UAAUAUACAAAUUAUGUGGCUUUAUUAAAA .................(-------(..((((((((((((.((((--.............-)-)))))))))....((-(-(--------......))))...))))))..))....... ( -19.52) >DroEre_CAF1 20987 98 + 1 UUUCCAUUCUCAAUCGCA-------A-UCCACAUGUCGUGACAAA--AACUCCCAUCUCA-U-GCGCACGACUUUCCU-G-U--------AUAUAUCCAAAUUAUGUGGCAUUAAUAAAA .................(-------(-(((((((((((((.....--.............-.-...)))))).....(-(-.--------.......))....)))))).)))....... ( -12.05) >DroYak_CAF1 37326 107 + 1 UUCCCAUUCUCAAUCACA-------A-UCCACAUUUCGUGGGGAA--AGCUUCCACGACUUU-UCGCACGACUUUUUU-G-UUAUUGUUUUUAUAUUAAAAUGAUGUGGCGUUAAUAAAA ..................-------.-.((((((((((((.((((--(((......).))))-)).))))).......-.-.....(((((......))))))))))))........... ( -21.00) >DroPer_CAF1 16110 99 + 1 UUGCCUUUCUCAGUCACA-------G-UCCGCAGGUCGUGACUGAUAAUUUCCCAAAGAA-UUUCGCAUGACAGUUUUAGAC--------UUCUAUCCAAAUUAUAUAC----AACACAA .(((.....((((((((.-------(-.((...)).)))))))))....(((.....)))-....))).........(((..--------..)))..............----....... ( -14.10) >consensus UUCCCAUUCUCAAUCACA_______A_UCCACAUGUCGUGACGAA__AAUUCCCAUCUCA_U_UCGCACGACUUUCUU_G_U________UUAUAUACAAAUUAUGUGGCGUUAAUAAAA ..................................((((((.(((...................)))))))))................................................ ( -5.74 = -6.33 + 0.58)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:52 2006