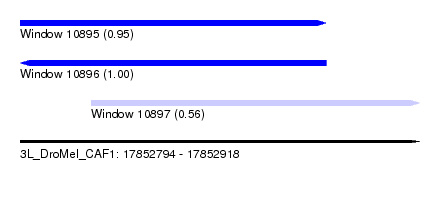
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,852,794 – 17,852,918 |
| Length | 124 |
| Max. P | 0.998759 |

| Location | 17,852,794 – 17,852,889 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 91.16 |
| Mean single sequence MFE | -20.32 |
| Consensus MFE | -17.72 |
| Energy contribution | -17.60 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.947870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17852794 95 + 23771897 GUUUUGCUUUUGCACGAUUAUUUCUGUUGCUGUUUUAUAGCAGUAAGCUAUAACUUUUACUACUAUUUUUCGCGCACGCUUCGUGACGUCACAGU (((.(((....))).)))..........(((((.(((((((.....)))))))..................(((((((...)))).))).))))) ( -18.00) >DroSec_CAF1 18381 95 + 1 GUUUUGCUUUUGCGCGAUUAUUUCUGUUGCUGUAUUAUAGCAGUAAGCUAUAACUUUUACUACUAUUUUACGCGCACGCUUCGUGACGUCACAGU .....((...(((((((.....)).......(((..((((.((((((........)))))).))))..)))))))).))...(((....)))... ( -21.60) >DroSim_CAF1 22427 95 + 1 GUUUUGCUUUUGCACGAUUAUUUCUGUUGCUGUUUUAUAGCAGUAAGCUAUAAUUUUUACUACUAUUUUUCGCGCACGCUUCGUGACGUCACAGU (((.(((....))).)))..........(((((.(((((((.....)))))))..................(((((((...)))).))).))))) ( -18.10) >DroEre_CAF1 19901 95 + 1 GUUUUGCUUUUGCACAACUAUUUCUGUUGCUGUUUUAUAGCAGUAAGCUAUAACUUUUACUACUAUUUUUCGCGCACGCUCCGUGACGUCACAGU (((.(((....))).)))..........(((((.(((((((.....)))))))..................(((((((...)))).))).))))) ( -20.00) >DroYak_CAF1 36354 86 + 1 GCUUUGCUUUUGCAAA---------GUUGCUGUUUUAUAGCAGUAAGCUAUAACUUUUAGUACUAUUUUGCGCGCACGCUUAGUGACGUCACAGU (((((((....)))))---------)).(((((.(((((((.....)))))))......((((((....(((....))).)))).))...))))) ( -23.90) >consensus GUUUUGCUUUUGCACGAUUAUUUCUGUUGCUGUUUUAUAGCAGUAAGCUAUAACUUUUACUACUAUUUUUCGCGCACGCUUCGUGACGUCACAGU (((.(((....))).)))..........(((((.(((((((.....)))))))..................((((((.....))).))).))))) (-17.72 = -17.60 + -0.12)

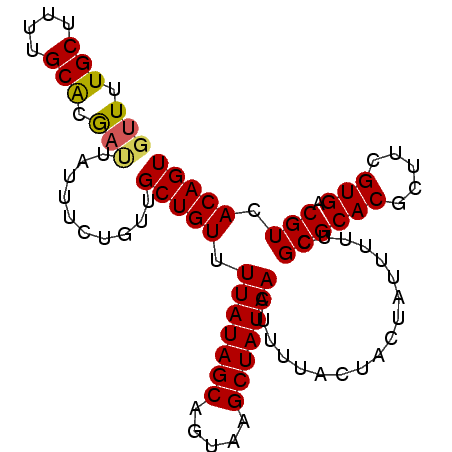
| Location | 17,852,794 – 17,852,889 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 91.16 |
| Mean single sequence MFE | -24.66 |
| Consensus MFE | -19.74 |
| Energy contribution | -20.14 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.79 |
| Structure conservation index | 0.80 |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.998759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17852794 95 - 23771897 ACUGUGACGUCACGAAGCGUGCGCGAAAAAUAGUAGUAAAAGUUAUAGCUUACUGCUAUAAAACAGCAACAGAAAUAAUCGUGCAAAAGCAAAAC ..((((....))))..((.(((((((...(((((((((..(((....))))))))))))...........(....)..)))))))...))..... ( -24.80) >DroSec_CAF1 18381 95 - 1 ACUGUGACGUCACGAAGCGUGCGCGUAAAAUAGUAGUAAAAGUUAUAGCUUACUGCUAUAAUACAGCAACAGAAAUAAUCGCGCAAAAGCAAAAC ..((((....))))..((.((((((((..(((((((((..(((....))))))))))))..))).......((.....)))))))...))..... ( -26.50) >DroSim_CAF1 22427 95 - 1 ACUGUGACGUCACGAAGCGUGCGCGAAAAAUAGUAGUAAAAAUUAUAGCUUACUGCUAUAAAACAGCAACAGAAAUAAUCGUGCAAAAGCAAAAC ..((((....))))..((.(((((((...((((((((((..........))))))))))...........(....)..)))))))...))..... ( -23.90) >DroEre_CAF1 19901 95 - 1 ACUGUGACGUCACGGAGCGUGCGCGAAAAAUAGUAGUAAAAGUUAUAGCUUACUGCUAUAAAACAGCAACAGAAAUAGUUGUGCAAAAGCAAAAC .(((((....))))).((.(((.......(((((((((..(((....))))))))))))......((((((....).))))))))...))..... ( -26.50) >DroYak_CAF1 36354 86 - 1 ACUGUGACGUCACUAAGCGUGCGCGCAAAAUAGUACUAAAAGUUAUAGCUUACUGCUAUAAAACAGCAAC---------UUUGCAAAAGCAAAGC .((((...((.((((.(((....)))....))))))......(((((((.....))))))).))))...(---------(((((....)))))). ( -21.60) >consensus ACUGUGACGUCACGAAGCGUGCGCGAAAAAUAGUAGUAAAAGUUAUAGCUUACUGCUAUAAAACAGCAACAGAAAUAAUCGUGCAAAAGCAAAAC ...(((....)))...((.(((((((......((.((.....(((((((.....)))))))....)).))........)))))))...))..... (-19.74 = -20.14 + 0.40)


| Location | 17,852,816 – 17,852,918 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 93.39 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -21.20 |
| Energy contribution | -22.64 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17852816 102 + 23771897 UCUGUUGCUGUUUUAUAGCAGUAAGCUAUAACUUUUACUACUAUUUUUCGCGCACGCUUCGUGACGUCACAGUCGUCGGAGUCGGCGAAAACGUUGUCCACU ....((((((((....))))))))....................(((((((.(..((((((((((......)))).)))))).))))))))........... ( -26.30) >DroSec_CAF1 18403 97 + 1 UCUGUUGCUGUAUUAUAGCAGUAAGCUAUAACUUUUACUACUAUUUUACGCGCACGCUUCGUGACGUCACAGUCGUCGGAGUCG----AAUCGUUGU-CACU .....(((((((..((((.((((((........)))))).))))..)))).)))(((((((((((......)))).))))).))----.........-.... ( -25.60) >DroSim_CAF1 22449 101 + 1 UCUGUUGCUGUUUUAUAGCAGUAAGCUAUAAUUUUUACUACUAUUUUUCGCGCACGCUUCGUGACGUCACAGUCGUCGGAGUCGGCGAAAACGUUGU-CACU ....((((((((....))))))))....................(((((((.(..((((((((((......)))).)))))).))))))))......-.... ( -26.30) >DroEre_CAF1 19923 101 + 1 UCUGUUGCUGUUUUAUAGCAGUAAGCUAUAACUUUUACUACUAUUUUUCGCGCACGCUCCGUGACGUCACAGUCGUCGGAGUAGGCGAAAACGUUGU-CACU ....((((((((....))))))))....................(((((((.(..((((((((((......)))).)))))).))))))))......-.... ( -29.20) >DroYak_CAF1 36370 98 + 1 ---GUUGCUGUUUUAUAGCAGUAAGCUAUAACUUUUAGUACUAUUUUGCGCGCACGCUUAGUGACGUCACAGUCGUCGGAGUCGGCGAAAACGUUGU-CACU ---..(((((..(((((((.....)))))))....))))).......(((....)))..(((((((......((((((....))))))......)))-)))) ( -28.20) >consensus UCUGUUGCUGUUUUAUAGCAGUAAGCUAUAACUUUUACUACUAUUUUUCGCGCACGCUUCGUGACGUCACAGUCGUCGGAGUCGGCGAAAACGUUGU_CACU ....((((((((....))))))))....................(((((((....((((((((((......)))).))))))..)))))))........... (-21.20 = -22.64 + 1.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:50 2006