| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
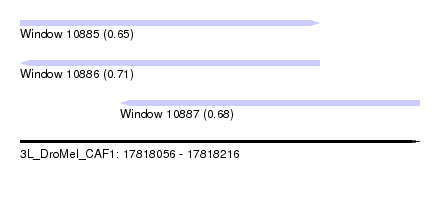
| Location | 17,818,056 – 17,818,216 |
| Length | 160 |
| Max. P | 0.712511 |

| Location | 17,818,056 – 17,818,176 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.33 |
| Mean single sequence MFE | -39.20 |
| Consensus MFE | -21.05 |
| Energy contribution | -21.67 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17818056 120 + 23771897 GCCAUAGGCCAGGAAGAUUGCCAGGCCUAUGGCGAUCCAUAUACUGAAGCGAAUCCAUGUGAGAAUGUCCAACUUGAUCAUCAGGUAAAUGUUGAUCAAAAUGGAAAUGCCGGGUAGCCA ((((((((((.((.......)).)))))))))).((((.................(((......)))((((..(((((((.((......)).)))))))..))))......))))..... ( -40.40) >DroPse_CAF1 148 120 + 1 GCCGUAGGCCAGAAAGAUCGCCAGACCUAUGGUGAGCCAAAUACAAAAACGCACCCAGGUGAGGAUGUCCAAUUUGAUCAUCAGGUAUAUGUUGAUCAAUAUGGAUAUGCCUGGGAGCCA ......(((........((((((......)))))))))............((.(((((((....(((((((..(((((((.((......)).)))))))..)))))))))))))).)).. ( -43.30) >DroEre_CAF1 153 120 + 1 GCCGUAGGCCAGAAAGAUAGUCAGGCCUAUUGCGAUCCAUAUGCUAAAGCGCACCCAUGUGAGAAUAUCCAACUUGAUCAUCAGGUAAAUGUUAAUCAGAAUGGAAAUGCCCGGUAGCCA ((.(((((((.((.......)).))))))).)).........((((..(.(((.((((.(((.(((((...(((((.....)))))..)))))..)))..))))...))).)..)))).. ( -33.60) >DroWil_CAF1 2637 120 + 1 GCUAUAGGAGAAAAAGAUGGCCAAACCGAUGGCAAUCCAAAUGCUAAAGCGCACCCAUGUUAGGAUAUCCAAUUUGAUCAUUAAGUAUAUAUUGAUCAGGAUGGAUAUGCCUGGUAGCCA (((((.((.......(((.((((......)))).)))....(((......))).))....(((((((((((.((((((((.((......)).)))))))).))))))).))))))))).. ( -37.20) >DroAna_CAF1 151 120 + 1 GCCGUAGGACAGGAAGAUGGACAGGCCGAUGGCCAGCCAUAUGCUGAAGCGCACCCAGGUGAGAAUGUCGAGCUUGAUCAUCAGGUAGAUGUUGAUCAGAAUGGAAAUGCCCGGCAGCCA ((((.....(((.(..((((...((((...))))..)))).).)))....(((.(((..(((.((((((..(((((.....))))).))))))..)))...)))...))).))))..... ( -36.40) >DroPer_CAF1 163 120 + 1 GCCGUUGGCCAUGAAGAUCGCCAGACCUAUGGUGAGCCAAAUACAAAAACGCACCCAGGUGAGGAUGUCCAAUUUGAUCAUCAGGUAUAUGUUGAUCAAUACGGAUACGCCUGGGAGCCA ....(((((........((((((......)))))))))))..........((.((((((((....(((((...(((((((.((......)).)))))))...))))))))))))).)).. ( -44.30) >consensus GCCGUAGGCCAGAAAGAUCGCCAGACCUAUGGCGAGCCAAAUACUAAAGCGCACCCAGGUGAGAAUGUCCAACUUGAUCAUCAGGUAUAUGUUGAUCAAAAUGGAAAUGCCUGGUAGCCA ......(((........((((((......)))))).((...........(.(((....))).)....((((..(((((((.((......)).)))))))..)))).......))..))). (-21.05 = -21.67 + 0.62)



| Location | 17,818,056 – 17,818,176 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.33 |
| Mean single sequence MFE | -35.88 |
| Consensus MFE | -23.86 |
| Energy contribution | -23.95 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.712511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17818056 120 - 23771897 UGGCUACCCGGCAUUUCCAUUUUGAUCAACAUUUACCUGAUGAUCAAGUUGGACAUUCUCACAUGGAUUCGCUUCAGUAUAUGGAUCGCCAUAGGCCUGGCAAUCUUCCUGGCCUAUGGC ..(((....)))...((((.((((((((.((......)).)))))))).))))............((((((..........))))))((((((((((.((.......)).)))))))))) ( -39.80) >DroPse_CAF1 148 120 - 1 UGGCUCCCAGGCAUAUCCAUAUUGAUCAACAUAUACCUGAUGAUCAAAUUGGACAUCCUCACCUGGGUGCGUUUUUGUAUUUGGCUCACCAUAGGUCUGGCGAUCUUUCUGGCCUACGGC ..((.((((((.((.((((..(((((((.((......)).)))))))..)))).)).....)))))).))....(((((...(((.((....(((((....)))))...)))))))))). ( -36.20) >DroEre_CAF1 153 120 - 1 UGGCUACCGGGCAUUUCCAUUCUGAUUAACAUUUACCUGAUGAUCAAGUUGGAUAUUCUCACAUGGGUGCGCUUUAGCAUAUGGAUCGCAAUAGGCCUGACUAUCUUUCUGGCCUACGGC ..((((..(.(((..((((...((((((.((......)).))))))...))))....(((....)))))).)..)))).........((..((((((.((.......)).))))))..)) ( -31.90) >DroWil_CAF1 2637 120 - 1 UGGCUACCAGGCAUAUCCAUCCUGAUCAAUAUAUACUUAAUGAUCAAAUUGGAUAUCCUAACAUGGGUGCGCUUUAGCAUUUGGAUUGCCAUCGGUUUGGCCAUCUUUUUCUCCUAUAGC ..((((..(((.(((((((...((((((.((......)).))))))...))))))))))....(((((((......)))...((((.((((......)))).))))......)))))))) ( -35.30) >DroAna_CAF1 151 120 - 1 UGGCUGCCGGGCAUUUCCAUUCUGAUCAACAUCUACCUGAUGAUCAAGCUCGACAUUCUCACCUGGGUGCGCUUCAGCAUAUGGCUGGCCAUCGGCCUGUCCAUCUUCCUGUCCUACGGC ..((((..(((((.........((((((.((......)).)))))).(((((.(((((......)))))))....)))..(((((.((((...)))).).)))).....)))))..)))) ( -33.90) >DroPer_CAF1 163 120 - 1 UGGCUCCCAGGCGUAUCCGUAUUGAUCAACAUAUACCUGAUGAUCAAAUUGGACAUCCUCACCUGGGUGCGUUUUUGUAUUUGGCUCACCAUAGGUCUGGCGAUCUUCAUGGCCAACGGC ..((.((((((.(..((((..(((((((.((......)).)))))))..))))......).)))))).))..........(((((.((....(((((....)))))...))))))).... ( -38.20) >consensus UGGCUACCAGGCAUAUCCAUUCUGAUCAACAUAUACCUGAUGAUCAAAUUGGACAUCCUCACAUGGGUGCGCUUUAGCAUAUGGAUCGCCAUAGGCCUGGCCAUCUUCCUGGCCUACGGC .(((((((((((...((((...((((((.((......)).))))))...))))(((((......)))))............(((....)))...)))))).........)))))...... (-23.86 = -23.95 + 0.09)
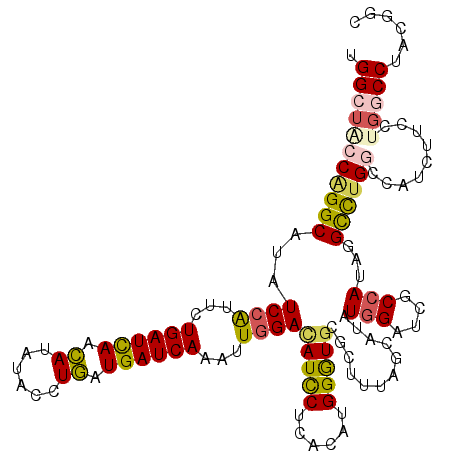
| Location | 17,818,096 – 17,818,216 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.17 |
| Mean single sequence MFE | -35.55 |
| Consensus MFE | -20.26 |
| Energy contribution | -20.02 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17818096 120 - 23771897 AUCAGGGGUGAAAUUGUCGUUCAAGGUGCCCUUGGUUCCCUGGCUACCCGGCAUUUCCAUUUUGAUCAACAUUUACCUGAUGAUCAAGUUGGACAUUCUCACAUGGAUUCGCUUCAGUAU ....((((((((..(((((.....((((((...((...)).))).))))))))..((((.((((((((.((......)).)))))))).))))..............))))))))..... ( -33.90) >DroPse_CAF1 188 120 - 1 AUCCGCAGUGAAUCUCUCCUUUAAGGUGCCCUUGGUUCCUUGGCUCCCAGGCAUAUCCAUAUUGAUCAACAUAUACCUGAUGAUCAAAUUGGACAUCCUCACCUGGGUGCGUUUUUGUAU ....((((.............(((((.(((...))).)))))((.((((((.((.((((..(((((((.((......)).)))))))..)))).)).....)))))).))....)))).. ( -34.40) >DroEre_CAF1 193 120 - 1 AUCAGGGGUGAAAUUGUCGUUCAAGGUUCCCUUGGUACCCUGGCUACCGGGCAUUUCCAUUCUGAUUAACAUUUACCUGAUGAUCAAGUUGGAUAUUCUCACAUGGGUGCGCUUUAGCAU ..((((((.((..(((.....)))..))))))))(((((((((((....)))...((((...((((((.((......)).))))))...))))........)).))))))((....)).. ( -34.90) >DroWil_CAF1 2677 120 - 1 UUCAGGUGUGAAGCUCUCAUUCAAAGUUCCUCUAGUACCCUGGCUACCAGGCAUAUCCAUCCUGAUCAAUAUAUACUUAAUGAUCAAAUUGGAUAUCCUAACAUGGGUGCGCUUUAGCAU .....(((((((((..........((.....)).((((((((......(((.(((((((...((((((.((......)).))))))...))))))))))..)).)))))))))))).))) ( -34.60) >DroAna_CAF1 191 120 - 1 GUCGGGAGUUAAGCUGUCCUUCAAGGUGCCAUUGGUACCAUGGCUGCCGGGCAUUUCCAUUCUGAUCAACAUCUACCUGAUGAUCAAGCUCGACAUUCUCACCUGGGUGCGCUUCAGCAU ...((((((..((((((((.....((((((((.(....)))))).)))))))..........((((((.((......)).))))))))))..))..))))......((((......)))) ( -34.80) >DroPer_CAF1 203 120 - 1 AUCCGCAGGGAAUCUCUGCUUUAAGGUGCCCUUGGUUCCUUGGCUCCCAGGCGUAUCCGUAUUGAUCAACAUAUACCUGAUGAUCAAAUUGGACAUCCUCACCUGGGUGCGUUUUUGUAU ....((((((...))))))..(((((.(((...))).)))))((.((((((.(..((((..(((((((.((......)).)))))))..))))......).)))))).)).......... ( -40.70) >consensus AUCAGGAGUGAAACUCUCCUUCAAGGUGCCCUUGGUACCCUGGCUACCAGGCAUAUCCAUUCUGAUCAACAUAUACCUGAUGAUCAAAUUGGACAUCCUCACAUGGGUGCGCUUUAGCAU ....((((((...............(((((..((((......))))...))))).((((...((((((.((......)).))))))...))))(((((......)))))))))))..... (-20.26 = -20.02 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:40 2006