
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,808,783 – 17,808,910 |
| Length | 127 |
| Max. P | 0.939828 |

| Location | 17,808,783 – 17,808,875 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.45 |
| Mean single sequence MFE | -23.07 |
| Consensus MFE | -12.95 |
| Energy contribution | -13.71 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17808783 92 - 23771897 AAUUUGGCGCCAAACACUUGUUAGCACUCUGUUGACUUUAACAUGGCGCAUUAACAGCUACGCUGAAAU----------------------CUGACAGA-UAGUAACAUAGCCUU .(((((((((((.......((((((.....)))))).......)))))).....((((...))))....----------------------....))))-).............. ( -23.54) >DroSec_CAF1 1710 92 - 1 AAUUUGGCGCCAAACACUUGUUAGCACUCUGUUGACUUUAACAGGGCGCACUAACAGCUCCCCUGAAAU----------------------CUGACAGA-UAGUAACAUAGCCUU .....(((((..(((....))).))..(((((((..(((..(((((.((.......))..)))))))).----------------------.)))))))-..........))).. ( -21.10) >DroSim_CAF1 1714 92 - 1 AAUUUGGCGCCAAACACUUGUUAGCACUCUGUUGACUUUAACAGGGCGCACUAACAGCUCCGCUGAAAU----------------------CUGACAGA-UAGUAACAUAGCCUU .....(((.......((((((((((.((((((((....)))))))).)).....((((...))))....----------------------.)))))..-.)))......))).. ( -23.32) >DroEre_CAF1 1630 92 - 1 AAUCUGCCGCCAAACACUUGUUAGCACUCUGUUGACUUUAACAUGGCGUCCUAACAGCAACGCUGAAAU----------------------CUGACAGC-UACUAGCAUAGCCUU ....((((((((.......((((((.....)))))).......))))).............((((....----------------------....))))-.....)))....... ( -19.74) >DroYak_CAF1 1643 92 - 1 AAUCUGGCGCCAAACACUUGUUAGCACUCUGUUGACCUUAACAGGGCGCCCUAACAGCUACGCUGAAAU----------------------CUGACAGA-UACUAACAUAGCCAU .((((((((((........((((((.....))))))........))))).....((((...))))....----------------------....))))-).............. ( -24.09) >DroAna_CAF1 1401 112 - 1 AAGUUUGCGCCAAACACCUGUUAUCCAACACCAGUGCUUAACAGAGUGC--UAACAU-AACAGUGAAGUAGUUAACAGAAUGGCUCAGUUUUUGACGUACUGCUAACAGUGCGCU ....(((.((((.....((((((....((...((..(((....)))..)--)..(((-....)))..))...))))))..)))).))).......(((((((....))))))).. ( -26.60) >consensus AAUUUGGCGCCAAACACUUGUUAGCACUCUGUUGACUUUAACAGGGCGCACUAACAGCUACGCUGAAAU______________________CUGACAGA_UACUAACAUAGCCUU ......(((((........((((((.....))))))........))))).....(((.....))).................................................. (-12.95 = -13.71 + 0.75)



| Location | 17,808,806 – 17,808,910 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 90.87 |
| Mean single sequence MFE | -26.13 |
| Consensus MFE | -21.53 |
| Energy contribution | -21.65 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17808806 104 - 23771897 ACAGAAGUUCUGCUUCCAAGGAGCCCUUAAAUUCAAAUUUGGCGCCAAACACUUGUUAGCACUCUGUUGACUUUAACAUGGCGCAUUAACAGCUACGCUGAAAU ...(((.....((((.....)))).......))).......((((((.......((((((.....)))))).......)))))).....((((...)))).... ( -25.54) >DroSec_CAF1 1733 104 - 1 ACAGAAGUUCUGCUUCCAAGGAGCCCUUAAAUUCAAAUUUGGCGCCAAACACUUGUUAGCACUCUGUUGACUUUAACAGGGCGCACUAACAGCUCCCCUGAAAU ...((((.....))))...(((((...(((((....)))))(((((........((((((.....))))))........))))).......)))))........ ( -27.49) >DroSim_CAF1 1737 104 - 1 ACAGAAGUUCUGCUCCCAAGGAGCCCUUAAAUUCAAAUUUGGCGCCAAACACUUGUUAGCACUCUGUUGACUUUAACAGGGCGCACUAACAGCUCCGCUGAAAU ...(((.....(((((...))))).......))).......(((((........((((((.....))))))........))))).....((((...)))).... ( -27.69) >DroEre_CAF1 1653 104 - 1 AACGAACUUCUGCGUUCAAGAAGCCUUUAAAUUCAAAUCUGCCGCCAAACACUUGUUAGCACUCUGUUGACUUUAACAUGGCGUCCUAACAGCAACGCUGAAAU .......(((.((((((..(((.........)))........(((((.......((((((.....)))))).......)))))........).))))).))).. ( -21.04) >DroYak_CAF1 1666 104 - 1 AAUGAAGUUCUGCUUCCGAGGAGCCUUUGAAUUCAAAUCUGGCGCCAAACACUUGUUAGCACUCUGUUGACCUUAACAGGGCGCCCUAACAGCUACGCUGAAAU ..((((.(((.((((.....))))....))))))).....((((((........((((((.....))))))........))))))....((((...)))).... ( -28.89) >consensus ACAGAAGUUCUGCUUCCAAGGAGCCCUUAAAUUCAAAUUUGGCGCCAAACACUUGUUAGCACUCUGUUGACUUUAACAGGGCGCACUAACAGCUACGCUGAAAU ...(((.....(((((...))))).......))).......(((((........((((((.....))))))........))))).....((((...)))).... (-21.53 = -21.65 + 0.12)

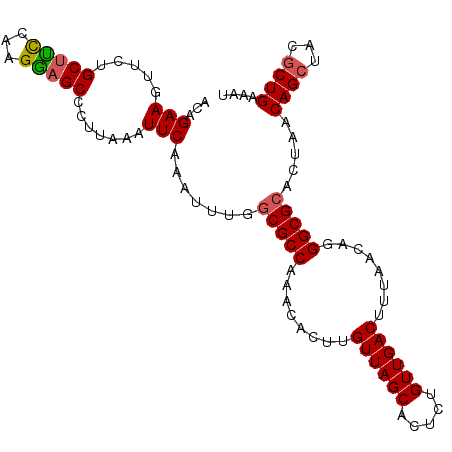
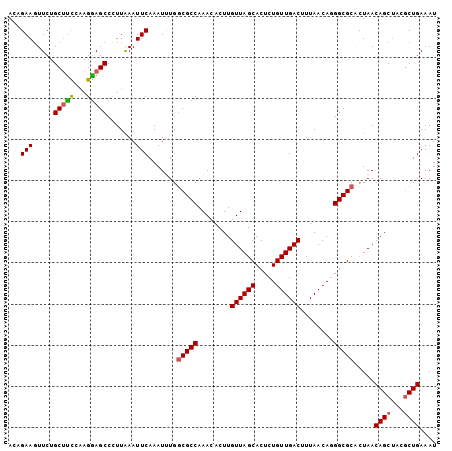
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:37 2006