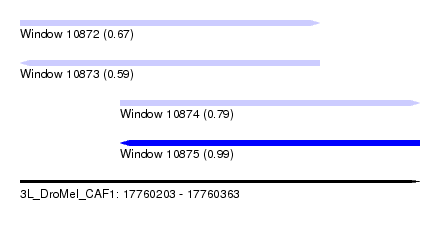
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,760,203 – 17,760,363 |
| Length | 160 |
| Max. P | 0.986064 |

| Location | 17,760,203 – 17,760,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.17 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -20.18 |
| Energy contribution | -21.86 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17760203 120 + 23771897 AUCUCUUUUCUCAGCACCCCCCGAUUGGCGAGAAUUGAGGUCCUCCGCCCCAAAGCUCAUGAAAAUCUCUUCCAGACUCACUGAGCUUAAGCUGAAUCCCCGAACACCCAAAAUGUUCAU ..........(((((...........((((((.((....)).)).))))...((((((((((...(((.....))).))).)))))))..)))))......(((((.......))))).. ( -27.60) >DroSec_CAF1 7657 120 + 1 AUCUCGUUUCUCAGCACCUCCCGAGUGGCGAGUAUUGAGGUCCUCCGCCCCAAAGCUCAUGAAAAUCUCUUCCAGACUCACUGAGCUUAAACUGAAUCCCCGAACACCUAAAGUGUUCAU ..(((((..(((..........)))..)))))...((.(((.....))).))((((((((((...(((.....))).))).))))))).............((((((.....)))))).. ( -29.50) >DroSim_CAF1 11734 120 + 1 AUCUCGUUUCUCAGCACCUCCCGAGUGGCGAGUAUUGAGGUCCUCCGCCCCAAAGCUCAUGAAAAUCUCUUCCAGACUCGCUGAGCUUAAACUGAAUCCCCGAACACCUAAAGUGUUCAU .....((((((((((.((........)).((((.(((.(((.....))).))).)))).....................))))))...)))).........((((((.....)))))).. ( -29.40) >DroEre_CAF1 7691 120 + 1 AUCUCUUUUCUCAGCACCUCCCAUUUUGCGCGUAUUUAGGUCCUCCGCUCCCAAGGUCAUGAAAAUCUCCUCCAGACUCACUGAGCUUAUGCUGAAACCCCGCACACCCAACGUGUUCAU ..........((((((......................((....))((((....((((..((........))..))))....))))...))))))......(.((((.....)))).).. ( -20.50) >DroYak_CAF1 25253 120 + 1 AUCUCUUUUGUCAGCACCUCCCGUUUUGCGAGUAUCGGCGUCCUCCGCCCCAAAGCUCAUGAAAAUCUCCUCCAGGCUCACUGAGCUCAUGCUGAAACCCCGAACACCCAAAGUAUUCAU ..........((((((.......((((..((((...((((.....)))).....))))..))))..........(((((...)))))..))))))......(((.((.....)).))).. ( -24.00) >consensus AUCUCUUUUCUCAGCACCUCCCGAUUGGCGAGUAUUGAGGUCCUCCGCCCCAAAGCUCAUGAAAAUCUCUUCCAGACUCACUGAGCUUAAGCUGAAUCCCCGAACACCCAAAGUGUUCAU ..........(((((...........((((((.((....)).)).))))...((((((((((...(((.....))).))).)))))))..)))))......((((((.....)))))).. (-20.18 = -21.86 + 1.68)

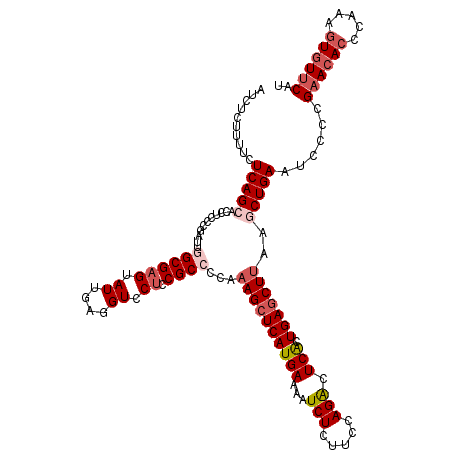
| Location | 17,760,203 – 17,760,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.17 |
| Mean single sequence MFE | -38.10 |
| Consensus MFE | -28.96 |
| Energy contribution | -29.32 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17760203 120 - 23771897 AUGAACAUUUUGGGUGUUCGGGGAUUCAGCUUAAGCUCAGUGAGUCUGGAAGAGAUUUUCAUGAGCUUUGGGGCGGAGGACCUCAAUUCUCGCCAAUCGGGGGGUGCUGAGAAAAGAGAU ..((((((.....))))))((((.(((.(((((((((((..((((((.....))))))...)))))))..)))).)))..))))...((((.....((((......)))).....)))). ( -39.70) >DroSec_CAF1 7657 120 - 1 AUGAACACUUUAGGUGUUCGGGGAUUCAGUUUAAGCUCAGUGAGUCUGGAAGAGAUUUUCAUGAGCUUUGGGGCGGAGGACCUCAAUACUCGCCACUCGGGAGGUGCUGAGAAACGAGAU ..((((((.....))))))((((.(((.(((((((((((..((((((.....))))))...)))))))..)))).)))..))))....((((...(((((......)))))...)))).. ( -41.60) >DroSim_CAF1 11734 120 - 1 AUGAACACUUUAGGUGUUCGGGGAUUCAGUUUAAGCUCAGCGAGUCUGGAAGAGAUUUUCAUGAGCUUUGGGGCGGAGGACCUCAAUACUCGCCACUCGGGAGGUGCUGAGAAACGAGAU ..((((((.....))))))((((.(((.(((((((((((..((((((.....))))))...)))))))..)))).)))..))))....((((...(((((......)))))...)))).. ( -40.90) >DroEre_CAF1 7691 120 - 1 AUGAACACGUUGGGUGUGCGGGGUUUCAGCAUAAGCUCAGUGAGUCUGGAGGAGAUUUUCAUGACCUUGGGAGCGGAGGACCUAAAUACGCGCAAAAUGGGAGGUGCUGAGAAAAGAGAU .((.((((.....)))).))...(((((((((...((((.((..(((.((((............)))).)))(((.............))).))...))))..)))))))))........ ( -30.92) >DroYak_CAF1 25253 120 - 1 AUGAAUACUUUGGGUGUUCGGGGUUUCAGCAUGAGCUCAGUGAGCCUGGAGGAGAUUUUCAUGAGCUUUGGGGCGGAGGACGCCGAUACUCGCAAAACGGGAGGUGCUGACAAAAGAGAU .(((((((.....))))))).....((((((((((((((.((((((....)).....)))))))))))...((((.....))))....(((.(.....).)))))))))).......... ( -37.40) >consensus AUGAACACUUUGGGUGUUCGGGGAUUCAGCUUAAGCUCAGUGAGUCUGGAAGAGAUUUUCAUGAGCUUUGGGGCGGAGGACCUCAAUACUCGCCAAUCGGGAGGUGCUGAGAAAAGAGAU ..((((((.....))))))(((..(((.((..(((((((..((((((.....))))))...)))))))....)).)))..))).....(((.....((((......)))).....))).. (-28.96 = -29.32 + 0.36)

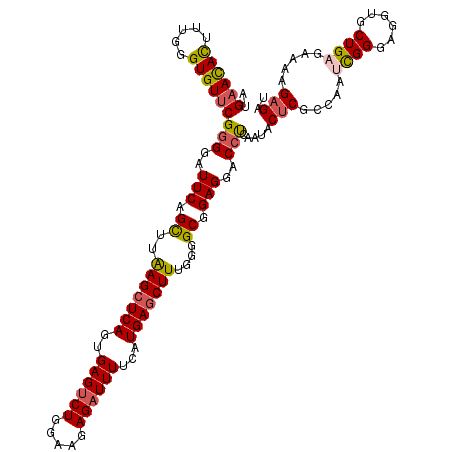

| Location | 17,760,243 – 17,760,363 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.50 |
| Mean single sequence MFE | -31.16 |
| Consensus MFE | -25.48 |
| Energy contribution | -25.84 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17760243 120 + 23771897 UCCUCCGCCCCAAAGCUCAUGAAAAUCUCUUCCAGACUCACUGAGCUUAAGCUGAAUCCCCGAACACCCAAAAUGUUCAUUUGGCUCUCCAAGUCACGGAAGAGCGAGGAAAAGUACUUU (((((.((......))..........(((((((.((((....(((....(((..(((....(((((.......))))))))..))))))..))))..))))))).))))).......... ( -31.10) >DroSec_CAF1 7697 120 + 1 UCCUCCGCCCCAAAGCUCAUGAAAAUCUCUUCCAGACUCACUGAGCUUAAACUGAAUCCCCGAACACCUAAAGUGUUCAUUUGGCUCUCCAAGUCACGGAAGAGCGAGGAAAAGGAAUUU (((((.((......))..........(((((((.((((....(((((.((...(......)((((((.....)))))).)).)))))....))))..))))))).))))).......... ( -33.60) >DroSim_CAF1 11774 120 + 1 UCCUCCGCCCCAAAGCUCAUGAAAAUCUCUUCCAGACUCGCUGAGCUUAAACUGAAUCCCCGAACACCUAAAGUGUUCAUUUGGCUCUCCAAGUCACGAAAGAGCGAGGAAAAGGAAUGU (((((((((...((((((((((...(((.....))).))).))))))).............((((((.....))))))....)))...........(....).).))))).......... ( -29.90) >DroEre_CAF1 7731 120 + 1 UCCUCCGCUCCCAAGGUCAUGAAAAUCUCCUCCAGACUCACUGAGCUUAUGCUGAAACCCCGCACACCCAACGUGUUCAUCUGGCGCUCCAAGUCGCGGAAGAGCGGGGAAUAGGAAUUG ...(((((((....((((..((........))..))))....))))...........((((((((((.....))))...(((..(((........)))..)))))))))....))).... ( -34.20) >DroYak_CAF1 25293 120 + 1 UCCUCCGCCCCAAAGCUCAUGAAAAUCUCCUCCAGGCUCACUGAGCUCAUGCUGAAACCCCGAACACCCAAAGUAUUCAUUUGGCGCUCCAAGUCGCGGAAGAGCGAGGAAAAGGAAUUU ...(((((......))...........((((((((.....))).((((.............(((.((.....)).))).(((.((((.....).))).))))))))))))...))).... ( -27.00) >consensus UCCUCCGCCCCAAAGCUCAUGAAAAUCUCUUCCAGACUCACUGAGCUUAAGCUGAAUCCCCGAACACCCAAAGUGUUCAUUUGGCUCUCCAAGUCACGGAAGAGCGAGGAAAAGGAAUUU (((((((((...((((((((((...(((.....))).))).))))))).............((((((.....))))))....)))...........(....).).))))).......... (-25.48 = -25.84 + 0.36)


| Location | 17,760,243 – 17,760,363 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.50 |
| Mean single sequence MFE | -42.40 |
| Consensus MFE | -36.86 |
| Energy contribution | -36.90 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17760243 120 - 23771897 AAAGUACUUUUCCUCGCUCUUCCGUGACUUGGAGAGCCAAAUGAACAUUUUGGGUGUUCGGGGAUUCAGCUUAAGCUCAGUGAGUCUGGAAGAGAUUUUCAUGAGCUUUGGGGCGGAGGA ..........((((((((((.(((.....)))))))).....((((((.....)))))))))))(((.(((((((((((..((((((.....))))))...)))))))..)))).))).. ( -42.00) >DroSec_CAF1 7697 120 - 1 AAAUUCCUUUUCCUCGCUCUUCCGUGACUUGGAGAGCCAAAUGAACACUUUAGGUGUUCGGGGAUUCAGUUUAAGCUCAGUGAGUCUGGAAGAGAUUUUCAUGAGCUUUGGGGCGGAGGA ....((((((((((((((((.(((.....)))))))).....((((((.....)))))))))))....(((((((((((..((((((.....))))))...)))))))..)))))))))) ( -44.80) >DroSim_CAF1 11774 120 - 1 ACAUUCCUUUUCCUCGCUCUUUCGUGACUUGGAGAGCCAAAUGAACACUUUAGGUGUUCGGGGAUUCAGUUUAAGCUCAGCGAGUCUGGAAGAGAUUUUCAUGAGCUUUGGGGCGGAGGA ....((((((((((((((((((........))))))).....((((((.....)))))))))))....(((((((((((..((((((.....))))))...)))))))..)))))))))) ( -41.50) >DroEre_CAF1 7731 120 - 1 CAAUUCCUAUUCCCCGCUCUUCCGCGACUUGGAGCGCCAGAUGAACACGUUGGGUGUGCGGGGUUUCAGCAUAAGCUCAGUGAGUCUGGAGGAGAUUUUCAUGACCUUGGGAGCGGAGGA ..........((((((((((((((.(((((.(((((((.((((....)))).)))((((.........))))..))))...))))))))))))...(..((......))..))))).))) ( -42.30) >DroYak_CAF1 25293 120 - 1 AAAUUCCUUUUCCUCGCUCUUCCGCGACUUGGAGCGCCAAAUGAAUACUUUGGGUGUUCGGGGUUUCAGCAUGAGCUCAGUGAGCCUGGAGGAGAUUUUCAUGAGCUUUGGGGCGGAGGA ....(((((..((((.((((((((((((((.(((((((.((........)).))))))).)))))...))....((((...))))..))))))).(((....)))....))))..))))) ( -41.40) >consensus AAAUUCCUUUUCCUCGCUCUUCCGUGACUUGGAGAGCCAAAUGAACACUUUGGGUGUUCGGGGAUUCAGCUUAAGCUCAGUGAGUCUGGAAGAGAUUUUCAUGAGCUUUGGGGCGGAGGA ..........((((((((((((((.....))))(((((...(((((((.....))))))).)).))).....(((((((..((((((.....))))))...))))))).))))))).))) (-36.86 = -36.90 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:28 2006