
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,754,833 – 17,755,073 |
| Length | 240 |
| Max. P | 0.999544 |

| Location | 17,754,833 – 17,754,953 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.72 |
| Mean single sequence MFE | -22.45 |
| Consensus MFE | -18.78 |
| Energy contribution | -18.22 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17754833 120 - 23771897 UAUAGUUACACACAUAGUAUAUCUUUUGUGGCGCCCCAUCAUCAAUAUUGAAAAGUGCACUCGCAGGAGCAAAAACAGCAAGAAAAACAAUUGAAAAGCACAAUAAAGUGUGCAACGUGA ..........(((...((.(.((((((((.((.((...(((.......)))....(((....))))).))....)))).)))).).)).........(((((......)))))...))). ( -22.00) >DroSec_CAF1 777 117 - 1 UAUAGUUACACACAU---AUAUCUUUUGUGGCGCCCCAUCAUCAAUACUGAAAAGUGCACUCGCAGGAGCAAAAACAGCAAGAAAAACAAUUGAAAAGCACAAUAAAGUGUGCAACGUGA ..........(((..---...((((((((.((.((...........(((....)))((....)).)).))....)))).))))..............(((((......)))))...))). ( -22.70) >DroSim_CAF1 3962 117 - 1 UAUAGUUACACACAU---AUAUCUUUUGUGGCGCCCCAUCAUCAAUAUUGAAAAGUGCACUCGCAGGAGCAAAAACAGCAAGAAAAACAAUUGAAAAGCACAAUAAAGUGUGCAACGUGA ..........(((..---...((((((((.((.((...(((.......)))....(((....))))).))....)))).))))..............(((((......)))))...))). ( -21.90) >DroEre_CAF1 3510 116 - 1 UAUAGUUACAUA-AU---AUAUCUUUUGUGGCGCCCCAUCAUCAAUAUUGGAAAGUGCAAUUGCAAGAGCAAAAAUAACAAGAGAAACAAUUGAAAAGCACAAUACAGUGUGCAACGUGA .....((((...-..---...((((((((.((((.(((..........)))...))))..((((....)))).....))))))))............(((((......)))))...)))) ( -25.50) >DroYak_CAF1 20004 116 - 1 UAUAGUUACAUA-AU---AUAUCUUUUGUGGCGCCCCAUCAUCAAUAUUGGAAAGUGCACUUGCAAGCUUAAAAAUAACAAGAAAAACAAUUGAAAUGCACAAUAAAGUGUGCAACGUGA .....((((...-..---...((..((((.((((.(((..........)))...)))).((((...............))))....))))..))..((((((......))))))..)))) ( -20.16) >consensus UAUAGUUACACACAU___AUAUCUUUUGUGGCGCCCCAUCAUCAAUAUUGAAAAGUGCACUCGCAGGAGCAAAAACAGCAAGAAAAACAAUUGAAAAGCACAAUAAAGUGUGCAACGUGA ........(((...........((((((((((((....(((.......)))...))))...))))))))............................(((((......)))))...))). (-18.78 = -18.22 + -0.56)

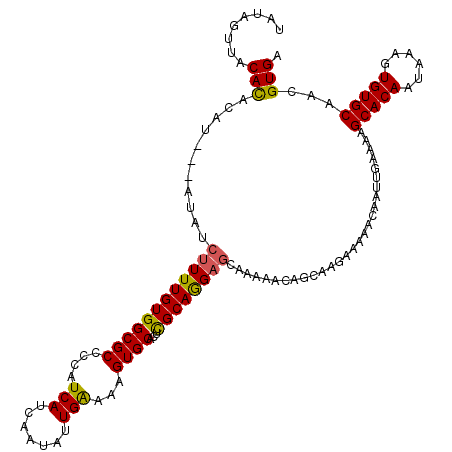
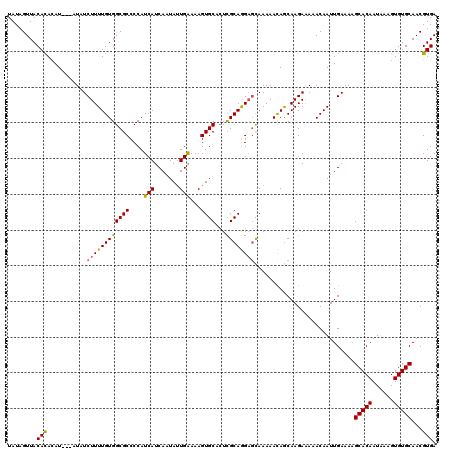
| Location | 17,754,913 – 17,755,033 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -21.36 |
| Consensus MFE | -19.03 |
| Energy contribution | -19.03 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17754913 120 - 23771897 CUUCACGUUUCACAUUUCAAUUCAUUGAUUAAGCAAAUGCAUUCAAUUUUCGAUUGGCUCUUAAUCAAUGAAGCGUUAAAUAUAGUUACACACAUAGUAUAUCUUUUGUGGCGCCCCAUC ....................(((((((((((((.....((..((.......))...)).)))))))))))))((((((.....((.(((.......)))...))....))))))...... ( -23.20) >DroSec_CAF1 857 114 - 1 CUUCACGUUU---AUUUCAAUUCAUUGAUUAAGCAAAUGCAUUCAAUUUUCGAUUGGCUCUUAAUCAAUGAAGCGUUAAAUAUAGUUACACACAU---AUAUCUUUUGUGGCGCCCCAUC ..........---.......(((((((((((((.....((..((.......))...)).)))))))))))))((((((.(((((((.....)).)---))))......))))))...... ( -21.30) >DroSim_CAF1 4042 114 - 1 CUUCACGUUU---AUUUCAAUUCAUUGAUUAAGCAAAUGCAUUCAAUUUUCGAUUGGCUCUUAAUCAAUGAAGCGUUAAAUAUAGUUACACACAU---AUAUCUUUUGUGGCGCCCCAUC ..........---.......(((((((((((((.....((..((.......))...)).)))))))))))))((((((.(((((((.....)).)---))))......))))))...... ( -21.30) >DroEre_CAF1 3590 112 - 1 CUUUACGUUU---GUUUCAAUUCAUUGAUUUGGCAAAAGCGUUCAAUUUUCGAUUGGC-CUUAAUCAAUGAUGCGUUAAAUAUAGUUACAUA-AU---AUAUCUUUUGUGGCGCCCCAUC ....((((..---........(((((((((.(((.....((.........))....))-)..))))))))).))))........((((((..-..---........))))))........ ( -20.40) >DroYak_CAF1 20084 113 - 1 CUUCACGUUU---AUUUCAAUUCAUUGAUUAGGCAAAUGCGUUCAAUUUUCGAUUGGCUCUUGAUCAAUGAAGCGUUAAAUAUAGUUACAUA-AU---AUAUCUUUUGUGGCGCCCCAUC ......((((---(...(..(((((((((((((.....((...(((((...))))))).)))))))))))))..).)))))...((((((..-..---........))))))........ ( -20.60) >consensus CUUCACGUUU___AUUUCAAUUCAUUGAUUAAGCAAAUGCAUUCAAUUUUCGAUUGGCUCUUAAUCAAUGAAGCGUUAAAUAUAGUUACACACAU___AUAUCUUUUGUGGCGCCCCAUC ....................(((((((((((((.....((..((.......))...)).)))))))))))))((((((.....((.................))....))))))...... (-19.03 = -19.03 + -0.00)

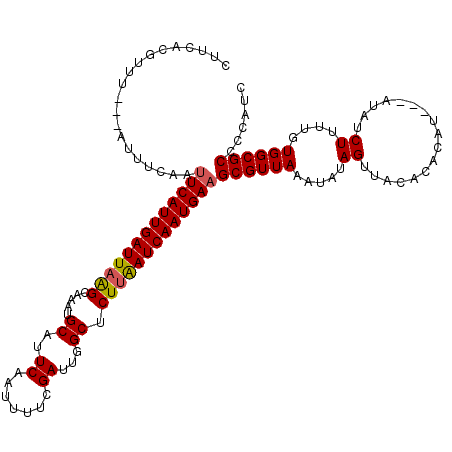
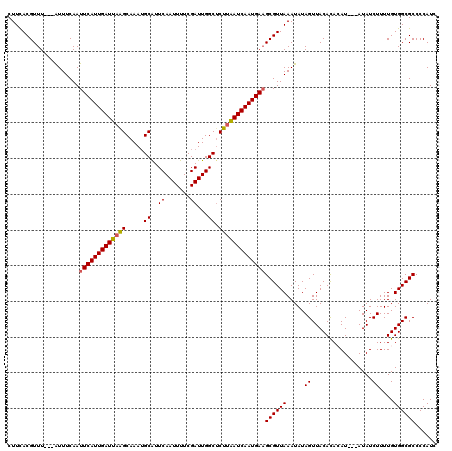
| Location | 17,754,953 – 17,755,073 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.20 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -21.38 |
| Energy contribution | -22.98 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17754953 120 + 23771897 UUUAACGCUUCAUUGAUUAAGAGCCAAUCGAAAAUUGAAUGCAUUUGCUUAAUCAAUGAAUUGAAAUGUGAAACGUGAAGCCAGCAACAAAGAAUGGGCUGGCUGUUUGUGAAAUGGGCU ......(((((((((((((((...((.((((...)))).))......))))))))))))........((...(((...(((((((..((.....)).)))))))...)))...)).))). ( -30.80) >DroSec_CAF1 894 117 + 1 UUUAACGCUUCAUUGAUUAAGAGCCAAUCGAAAAUUGAAUGCAUUUGCUUAAUCAAUGAAUUGAAAU---AAACGUGAAGCCAGCAACAAAGAAUGGGCUGGCUGUUUGUGAAAUGCGCU .....((((((((((((((((...((.((((...)))).))......))))))))))))).....((---((((....(((((((..((.....)).))))))))))))).....))).. ( -32.50) >DroSim_CAF1 4079 117 + 1 UUUAACGCUUCAUUGAUUAAGAGCCAAUCGAAAAUUGAAUGCAUUUGCUUAAUCAAUGAAUUGAAAU---AAACGUGAAGCCAGCAACAAAGAAUGGGCUGGCUGUUUGUGAAAUGCGCU .....((((((((((((((((...((.((((...)))).))......))))))))))))).....((---((((....(((((((..((.....)).))))))))))))).....))).. ( -32.50) >DroEre_CAF1 3626 112 + 1 UUUAACGCAUCAUUGAUUAAG-GCCAAUCGAAAAUUGAACGCUUUUGCCAAAUCAAUGAAUUGAAAC---AAACGUAAAGCCCACAAUGGAGAAUGAGGUGGCU----GCGAAAUGCGCU .....(((((((((((((..(-((.((.((.........))..)).))).)))))))))........---...(((..((((.((.((.....))...))))))----)))...)))).. ( -24.20) >DroYak_CAF1 20120 113 + 1 UUUAACGCUUCAUUGAUCAAGAGCCAAUCGAAAAUUGAACGCAUUUGCCUAAUCAAUGAAUUGAAAU---AAACGUGAAGCCAACUAUAUGGAAUGAGGUGGCU----CUGAAAUUCGCU ................(((.((((((.((((..(((((..((....))....)))))...))))...---...(((....(((......))).)))...)))))----))))........ ( -21.00) >consensus UUUAACGCUUCAUUGAUUAAGAGCCAAUCGAAAAUUGAAUGCAUUUGCUUAAUCAAUGAAUUGAAAU___AAACGUGAAGCCAGCAACAAAGAAUGGGCUGGCUGUUUGUGAAAUGCGCU ......(((((((((((((((.((...((((...))))..)).....)))))))))))))..................(((((((............))))))).............)). (-21.38 = -22.98 + 1.60)

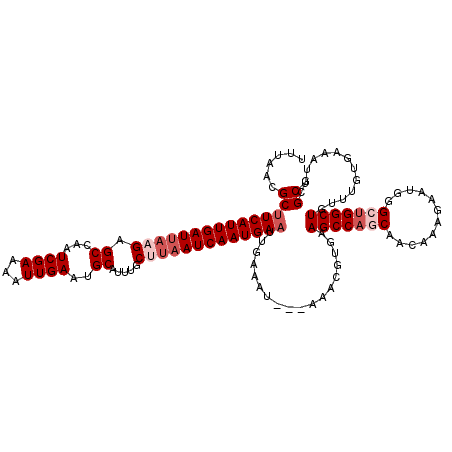
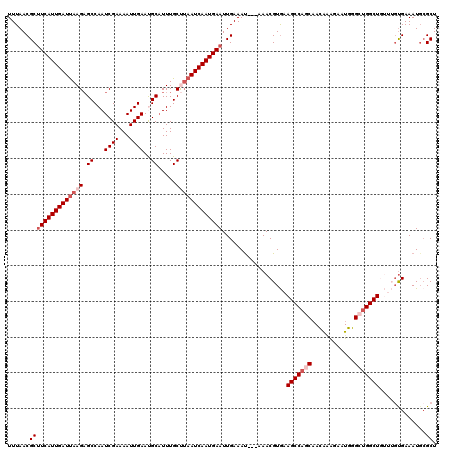
| Location | 17,754,953 – 17,755,073 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.20 |
| Mean single sequence MFE | -29.78 |
| Consensus MFE | -25.36 |
| Energy contribution | -26.36 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.71 |
| SVM RNA-class probability | 0.999544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17754953 120 - 23771897 AGCCCAUUUCACAAACAGCCAGCCCAUUCUUUGUUGCUGGCUUCACGUUUCACAUUUCAAUUCAUUGAUUAAGCAAAUGCAUUCAAUUUUCGAUUGGCUCUUAAUCAAUGAAGCGUUAAA .((.............(((((((.((.....))..)))))))..................(((((((((((((.....((..((.......))...)).)))))))))))))))...... ( -29.10) >DroSec_CAF1 894 117 - 1 AGCGCAUUUCACAAACAGCCAGCCCAUUCUUUGUUGCUGGCUUCACGUUU---AUUUCAAUUCAUUGAUUAAGCAAAUGCAUUCAAUUUUCGAUUGGCUCUUAAUCAAUGAAGCGUUAAA (((((.......(((((((((((.((.....))..)))))))....))))---.......(((((((((((((.....((..((.......))...)).))))))))))))))))))... ( -34.30) >DroSim_CAF1 4079 117 - 1 AGCGCAUUUCACAAACAGCCAGCCCAUUCUUUGUUGCUGGCUUCACGUUU---AUUUCAAUUCAUUGAUUAAGCAAAUGCAUUCAAUUUUCGAUUGGCUCUUAAUCAAUGAAGCGUUAAA (((((.......(((((((((((.((.....))..)))))))....))))---.......(((((((((((((.....((..((.......))...)).))))))))))))))))))... ( -34.30) >DroEre_CAF1 3626 112 - 1 AGCGCAUUUCGC----AGCCACCUCAUUCUCCAUUGUGGGCUUUACGUUU---GUUUCAAUUCAUUGAUUUGGCAAAAGCGUUCAAUUUUCGAUUGGC-CUUAAUCAAUGAUGCGUUAAA ((((((......----((((....((........))..))))........---........(((((((((.(((.....((.........))....))-)..)))))))))))))))... ( -26.90) >DroYak_CAF1 20120 113 - 1 AGCGAAUUUCAG----AGCCACCUCAUUCCAUAUAGUUGGCUUCACGUUU---AUUUCAAUUCAUUGAUUAGGCAAAUGCGUUCAAUUUUCGAUUGGCUCUUGAUCAAUGAAGCGUUAAA .((((((....(----(((((.((.((.....)))).))))))...))))---.......(((((((((((((.....((...(((((...))))))).)))))))))))))))...... ( -24.30) >consensus AGCGCAUUUCACAAACAGCCAGCCCAUUCUUUGUUGCUGGCUUCACGUUU___AUUUCAAUUCAUUGAUUAAGCAAAUGCAUUCAAUUUUCGAUUGGCUCUUAAUCAAUGAAGCGUUAAA (((((...........(((((((............)))))))..................(((((((((((((.....((..((.......))...)).))))))))))))))))))... (-25.36 = -26.36 + 1.00)

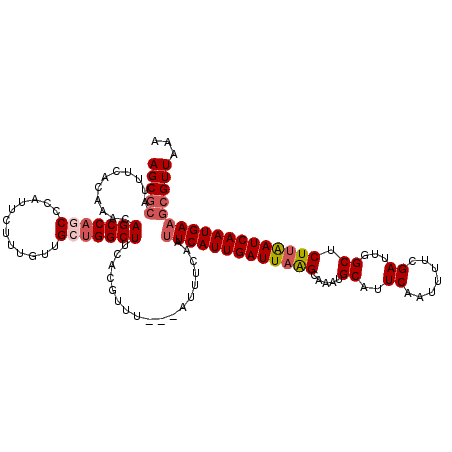
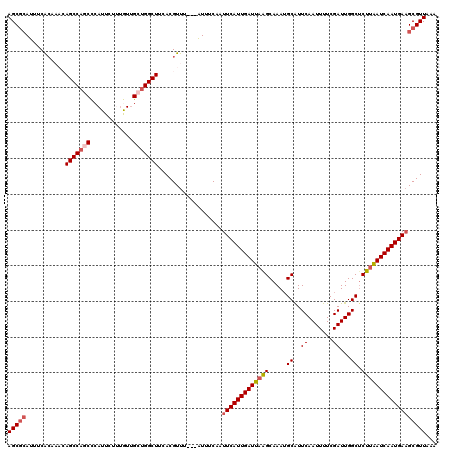
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:23 2006