| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
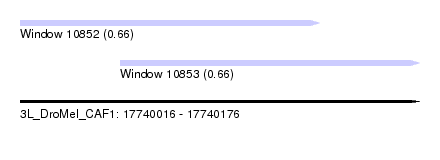
| Location | 17,740,016 – 17,740,176 |
| Length | 160 |
| Max. P | 0.662266 |

| Location | 17,740,016 – 17,740,136 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.93 |
| Mean single sequence MFE | -31.23 |
| Consensus MFE | -21.86 |
| Energy contribution | -22.80 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17740016 120 + 23771897 AAUAACAGUUAAAAAUGCCAUGAGAAUUCGAAUUAUUUCCAAAUGCAAUUACAGUGUUCAGUGGCGACUGUCCAUAAGUCAAGGUUCCCAAAUUGAACCCACUUUGGACAGGUGGUCUUA ...............((((((..((((.((((....)))....((......))).)))).)))))).(((((((.((((...(((((.......))))).)))))))))))......... ( -28.20) >DroSec_CAF1 21227 119 + 1 AAUAGCAGUUAAAAAUGCCAUGAGAAUUCGAAUUAUUUCGAAAUGCAAUUACAGUGUUCAAUGGCCACUGUCCAUA-GUCAAGGUUCCUAAUUUGAACCUACUUUGGACAGCUGGUCUUA ....(((........)))..((((..((((((....))))))..((.......)).))))..((((((((((((.(-((..((((((.......))))))))).))))))).)))))... ( -35.20) >DroSim_CAF1 24480 119 + 1 AAUAGCAGUUAAAAAUGCCAUGAGAAUUCGAAUUAUUUCGAAAUGCAAUUACAGUGUUCAGUGGCCACUGUCCAUA-GUCAAGGUUCCUAAAUUGAACCCACUUUGGACAGCUGGUCUUA ....(((........)))..((((..((((((....))))))..((.......)).))))..((((((((((((.(-((...(((((.......))))).))).))))))).)))))... ( -33.50) >DroEre_CAF1 35986 117 + 1 AAUAGCAGUUAAAAAUGUCAUGAGAAUACGAAUGAUACCGAAAUGCAAUUAGUGCGUUC-UUGGCCACUGUCUAUA-GUCAAGGUUCCUAAAUUGAACCCACUUUGG-CAGCUGGUCUAA ...............((((((..(....)..))))))..(((.((((.....)))))))-..((((((((((...(-((...(((((.......))))).)))..))-))).)))))... ( -28.00) >consensus AAUAGCAGUUAAAAAUGCCAUGAGAAUUCGAAUUAUUUCGAAAUGCAAUUACAGUGUUCAGUGGCCACUGUCCAUA_GUCAAGGUUCCUAAAUUGAACCCACUUUGGACAGCUGGUCUUA .............(((((..(((...((((((....))))))......)))..)))))....((((((((((((...((...(((((.......))))).))..))))))).)))))... (-21.86 = -22.80 + 0.94)



| Location | 17,740,056 – 17,740,176 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.05 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -22.81 |
| Energy contribution | -23.37 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17740056 120 + 23771897 AAAUGCAAUUACAGUGUUCAGUGGCGACUGUCCAUAAGUCAAGGUUCCCAAAUUGAACCCACUUUGGACAGGUGGUCUUACAAAUUAUUCACGGCAUAAUUACACACCAAGGCUUCAGGG ....((.......((((...(((.(..(((((((.((((...(((((.......))))).)))))))))))((((.............))))).)))....))))......))....... ( -28.94) >DroSec_CAF1 21267 119 + 1 AAAUGCAAUUACAGUGUUCAAUGGCCACUGUCCAUA-GUCAAGGUUCCUAAUUUGAACCUACUUUGGACAGCUGGUCUUACAAAUUAUUCACGGCAUAAUUACACACCAAGGCUUCUGGG ..((((........(((.....((((((((((((.(-((..((((((.......))))))))).))))))).)))))..)))...........)))).........(((.......))). ( -33.81) >DroSim_CAF1 24520 119 + 1 AAAUGCAAUUACAGUGUUCAGUGGCCACUGUCCAUA-GUCAAGGUUCCUAAAUUGAACCCACUUUGGACAGCUGGUCUUACAAAUUAUUCACUGCAUAAUUACACACCAAGGCUUCUGGA ...((.((((((((((......((((((((((((.(-((...(((((.......))))).))).))))))).)))))............)))))..))))).))..(((.......))). ( -32.67) >DroEre_CAF1 36026 117 + 1 AAAUGCAAUUAGUGCGUUC-UUGGCCACUGUCUAUA-GUCAAGGUUCCUAAAUUGAACCCACUUUGG-CAGCUGGUCUAACAAAUUAUUCACGGCAUAAUUACACACCAAGGCUUCUCGA ..((((...((((..(((.-..((((((((((...(-((...(((((.......))))).)))..))-))).))))).)))..))))......))))....................... ( -25.70) >consensus AAAUGCAAUUACAGUGUUCAGUGGCCACUGUCCAUA_GUCAAGGUUCCUAAAUUGAACCCACUUUGGACAGCUGGUCUUACAAAUUAUUCACGGCAUAAUUACACACCAAGGCUUCUGGA ....((.......((((.....((((((((((((...((...(((((.......))))).))..))))))).))))).....((((((.......))))))))))......))....... (-22.81 = -23.37 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:07 2006