
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,738,121 – 17,738,321 |
| Length | 200 |
| Max. P | 0.923702 |

| Location | 17,738,121 – 17,738,241 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -40.11 |
| Consensus MFE | -36.84 |
| Energy contribution | -37.02 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17738121 120 + 23771897 CAUUAUCCUUUUGGGCAACCGCCAAGGAUGUGGUGAUCCCAGUGGUCGCACCUCCACAUAUCAGCUUACCAAUAAGAUGACUCCUGUCCUGGUUUCGGGCGUCAAGUGGCAUUAUCGCCA ....((((((.(((....)))..))))))((((((((.(((((((........))))....................((((.((((.........)))).))))..))).)))))))).. ( -36.30) >DroSec_CAF1 19323 120 + 1 CAUUAUCCUUGUGGGCAACCGCCAAGGAUGUGGCGAUCCCAGUGGUCGCAUCUCCACAUAUCAGCUUACCAAUGAGAUGACUCCUCCCCUUGUUUCGGGUGUCAAGUGGCAUUAUCGCCA ...(((((((((((....))).))))))))((((((((((.((((........))))...(((.((((....)))).)))................)))((((....))))..))))))) ( -40.80) >DroSim_CAF1 22593 120 + 1 CAUUAUCCUUGUGGGCAACCGCCAAGGAUGUGGUGAUCCCAGUGGUCGCAUCUCCACAUAUCAGCUUACCAAUGCGAUGACUCCUGCCCUUGUUUCGGGUGUCAAGUGGCAUUAUCGCCA ...(((((((((((....))).))))))))(((((((...(((.(((((((....................))))))).)))..((((((((..........)))).))))..))))))) ( -42.55) >DroEre_CAF1 34095 119 + 1 CAUUAUCCUUGUGGGCAACCACCAAGGAUGUGGUGAUCCCAGUGGUCGCAUCUCCACAUAUCAGCUUACCAAUGAGAUGACUCCUGCCCU-GUUUCGGGCGUCAAGUGGCAUUAUCGACG ...(((((((((((....))).))))))))(((((((.(((.(((...((((((...................))))))...)).((((.-.....))))....).))).)))))))... ( -40.81) >consensus CAUUAUCCUUGUGGGCAACCGCCAAGGAUGUGGUGAUCCCAGUGGUCGCAUCUCCACAUAUCAGCUUACCAAUGAGAUGACUCCUGCCCUUGUUUCGGGCGUCAAGUGGCAUUAUCGCCA ...(((((((((((....))).))))))))(((((((.(((((((........))))....................((((.((((.........)))).))))..)))....))))))) (-36.84 = -37.02 + 0.19)

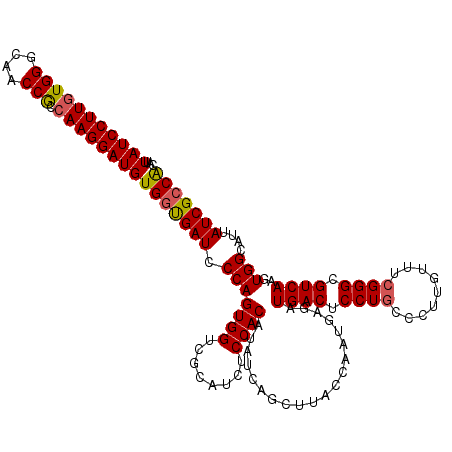

| Location | 17,738,161 – 17,738,281 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.75 |
| Mean single sequence MFE | -31.08 |
| Consensus MFE | -28.05 |
| Energy contribution | -28.55 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17738161 120 + 23771897 AGUGGUCGCACCUCCACAUAUCAGCUUACCAAUAAGAUGACUCCUGUCCUGGUUUCGGGCGUCAAGUGGCAUUAUCGCCAGAACAGUUGUCAUCCCCUUCGACAACAAUUAUUAUGCCAC .((((........)))).......((((....)))).((((.((((.........)))).)))).(((((((..((....))...((((((.........)))))).......))))))) ( -30.70) >DroSec_CAF1 19363 120 + 1 AGUGGUCGCAUCUCCACAUAUCAGCUUACCAAUGAGAUGACUCCUCCCCUUGUUUCGGGUGUCAAGUGGCAUUAUCGCCAGAACAGUUGUCAUCCCCUUCGACAACAAGUAUUAUGCCAC .((((........)))).......((((....)))).((((....(((........))).)))).(((((((..((....))...((((((.........)))))).......))))))) ( -28.90) >DroSim_CAF1 22633 120 + 1 AGUGGUCGCAUCUCCACAUAUCAGCUUACCAAUGCGAUGACUCCUGCCCUUGUUUCGGGUGUCAAGUGGCAUUAUCGCCAGAACAGUUGUCAUCCCCUUCGACAACAAUUAUUAUGCCAC .((((........))))......((........))..((((.((((.........)))).)))).(((((((..((....))...((((((.........)))))).......))))))) ( -31.60) >DroEre_CAF1 34135 119 + 1 AGUGGUCGCAUCUCCACAUAUCAGCUUACCAAUGAGAUGACUCCUGCCCU-GUUUCGGGCGUCAAGUGGCAUUAUCGACGGAACUGCUGUCAUGCCCUCCGACAACAAUUAUUAUGCCAC .(((((..((((((...................))))))......((((.-.....))))(((.((.(((((....(((((.....))))))))))))..)))............))))) ( -33.11) >consensus AGUGGUCGCAUCUCCACAUAUCAGCUUACCAAUGAGAUGACUCCUGCCCUUGUUUCGGGCGUCAAGUGGCAUUAUCGCCAGAACAGUUGUCAUCCCCUUCGACAACAAUUAUUAUGCCAC .((((........))))....................((((.((((.........)))).)))).(((((((..((....))...((((((.........)))))).......))))))) (-28.05 = -28.55 + 0.50)

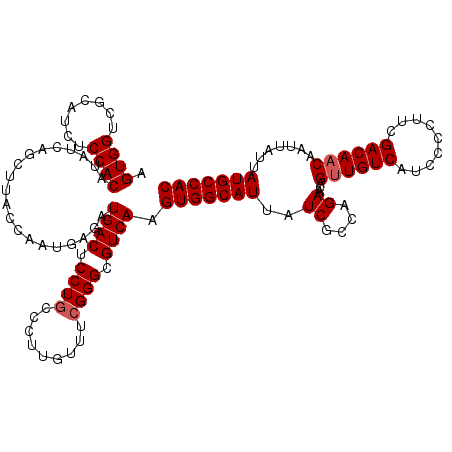
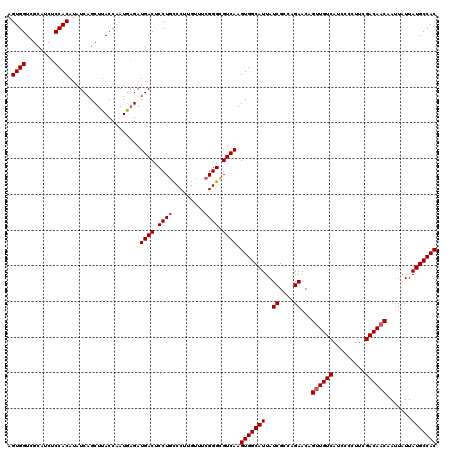
| Location | 17,738,201 – 17,738,321 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.03 |
| Mean single sequence MFE | -29.23 |
| Consensus MFE | -24.11 |
| Energy contribution | -24.80 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17738201 120 + 23771897 CUCCUGUCCUGGUUUCGGGCGUCAAGUGGCAUUAUCGCCAGAACAGUUGUCAUCCCCUUCGACAACAAUUAUUAUGCCACUUCUUGGCAUUUAAUUACAAUUUUCUAGGCAAUUCCAGUU .....((((.......))))(((....)))......(((((((..((((((.........))))))(((((..((((((.....)))))).)))))......)))).))).......... ( -30.10) >DroSec_CAF1 19403 120 + 1 CUCCUCCCCUUGUUUCGGGUGUCAAGUGGCAUUAUCGCCAGAACAGUUGUCAUCCCCUUCGACAACAAGUAUUAUGCCACUUCUUGCCAUUUAAUUACAAUUUUCUAGGCAAUUCCAGUU ...(.(((........))).)..(((((((((..((....))...((((((.........)))))).......))))))))).(((((...................)))))........ ( -26.01) >DroSim_CAF1 22673 120 + 1 CUCCUGCCCUUGUUUCGGGUGUCAAGUGGCAUUAUCGCCAGAACAGUUGUCAUCCCCUUCGACAACAAUUAUUAUGCCACUUCUUGCCAUUUAAUUACAAUUUUCUAGGCAAUUCCAGUU .....((((.......))))...(((((((((..((....))...((((((.........)))))).......))))))))).(((((...................)))))........ ( -27.91) >DroEre_CAF1 34175 119 + 1 CUCCUGCCCU-GUUUCGGGCGUCAAGUGGCAUUAUCGACGGAACUGCUGUCAUGCCCUCCGACAACAAUUAUUAUGCCACUUCUUGGCAUUUAAUUACAAUUUUCUAGGCAAUUCCAGUG ..(((((((.-.....))))(((.((.(((((....(((((.....))))))))))))..)))...(((((..((((((.....)))))).)))))..........)))........... ( -32.90) >consensus CUCCUGCCCUUGUUUCGGGCGUCAAGUGGCAUUAUCGCCAGAACAGUUGUCAUCCCCUUCGACAACAAUUAUUAUGCCACUUCUUGCCAUUUAAUUACAAUUUUCUAGGCAAUUCCAGUU ..(((((((.......))))((((((((((((..((....))...((((((.........)))))).......))))))))...))))..................)))........... (-24.11 = -24.80 + 0.69)

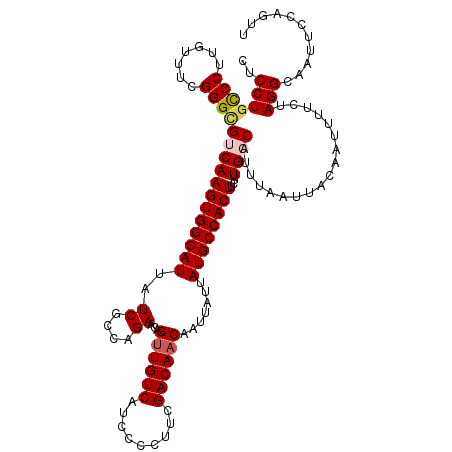

| Location | 17,738,201 – 17,738,321 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.03 |
| Mean single sequence MFE | -33.91 |
| Consensus MFE | -26.04 |
| Energy contribution | -27.10 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17738201 120 - 23771897 AACUGGAAUUGCCUAGAAAAUUGUAAUUAAAUGCCAAGAAGUGGCAUAAUAAUUGUUGUCGAAGGGGAUGACAACUGUUCUGGCGAUAAUGCCACUUGACGCCCGAAACCAGGACAGGAG ..((((......))))..............((((((.....)))))).....((((..((......))..))))(((((((((.......((........))......)))))))))... ( -30.62) >DroSec_CAF1 19403 120 - 1 AACUGGAAUUGCCUAGAAAAUUGUAAUUAAAUGGCAAGAAGUGGCAUAAUACUUGUUGUCGAAGGGGAUGACAACUGUUCUGGCGAUAAUGCCACUUGACACCCGAAACAAGGGGAGGAG ........(((((...................))))).(((((((((......((((((((((.((........)).))).))))))))))))))))....(((.......)))...... ( -32.51) >DroSim_CAF1 22673 120 - 1 AACUGGAAUUGCCUAGAAAAUUGUAAUUAAAUGGCAAGAAGUGGCAUAAUAAUUGUUGUCGAAGGGGAUGACAACUGUUCUGGCGAUAAUGCCACUUGACACCCGAAACAAGGGCAGGAG ..(((...(((((...................))))).(((((((((......((((((((((.((........)).))).))))))))))))))))....(((.......))))))... ( -34.11) >DroEre_CAF1 34175 119 - 1 CACUGGAAUUGCCUAGAAAAUUGUAAUUAAAUGCCAAGAAGUGGCAUAAUAAUUGUUGUCGGAGGGCAUGACAGCAGUUCCGUCGAUAAUGCCACUUGACGCCCGAAAC-AGGGCAGGAG ..((((......)))).................((...(((((((((......(((((.(((((.((......))..))))).))))))))))))))...((((.....-.)))).)).. ( -38.40) >consensus AACUGGAAUUGCCUAGAAAAUUGUAAUUAAAUGCCAAGAAGUGGCAUAAUAAUUGUUGUCGAAGGGGAUGACAACUGUUCUGGCGAUAAUGCCACUUGACACCCGAAACAAGGGCAGGAG ..((((......))))......................(((((((((......((((((((((.((........)).))).))))))))))))))))...((((.......))))..... (-26.04 = -27.10 + 1.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:04 2006