
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,735,340 – 17,735,710 |
| Length | 370 |
| Max. P | 0.999538 |

| Location | 17,735,340 – 17,735,439 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 93.63 |
| Mean single sequence MFE | -43.10 |
| Consensus MFE | -36.04 |
| Energy contribution | -36.32 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17735340 99 + 23771897 AAGGGUCGAGUGGCAGACAGUCGGACUGGGCCGCAUUUGCAGCUGCUCUUGGCCAUAGCCGCCCCCUUUGGGGCGUGGCCGUGUCUGAGCCAUAAAACC ...(((...(((((......((((((..((((((....((....))....(((....)))(((((....)))))))))))..)))))))))))...))) ( -42.70) >DroSec_CAF1 16462 99 + 1 AAGGGUCGAGUGGCAGACAGUCGGACUGGGCCGCGUUUGCAGCGGCUCUUGGCCAUAGCCGCCCCUUUGACGGCGUGGCCGUGUUUGGGCCAUAAAACC ...(((...(((((.((((........(((((((.......)))))))..((((((.(((((......).)))))))))).))))...)))))...))) ( -42.00) >DroSim_CAF1 19179 99 + 1 AAGGGUCGAGUGGCAGACAGUCGGACUGGGCCGCGUUUGCAGCUGCUCUUGGCCAUAGCCGCCCCUUUGGCGGCGUGGCCGUGUCUGGGCCAUAAAACC ...(((...(((((...(((.((....((((.((.......)).))))..((((((.((((((.....)))))))))))).)).))).)))))...))) ( -45.30) >DroEre_CAF1 31388 98 + 1 AAGGGUCGAGUGGCAGACAGUCGGACUGGGCCGCGUUUGCAGUGGCUCUUGGCCAUAGCCGCCCAUUUG-GGGCGUGGCCGUGUCUGGACCAUAAAACC ...((((((.((.....)).))((((..((((((....((.(((((.....))))).)).((((.....-))))))))))..)))).))))........ ( -45.00) >DroYak_CAF1 32006 98 + 1 AAGGGUCGAGUGGCAGACAGUCGGACUGGGCCGCAUUUGCAGCUGCUCCUGGCCAUAGCCGCCCUUUUU-GGGCGUGGCCGUGUCUGGGCCAUAAAACC ...(((...(((((...(((.((((...(((.((....)).)))..))).(((((....(((((.....-))))))))))..).))).)))))...))) ( -40.50) >consensus AAGGGUCGAGUGGCAGACAGUCGGACUGGGCCGCGUUUGCAGCUGCUCUUGGCCAUAGCCGCCCCUUUG_GGGCGUGGCCGUGUCUGGGCCAUAAAACC ...((((..(((((...(((.....))).))))).....(((..((...((((((....(((((......))))))))))).)))))))))........ (-36.04 = -36.32 + 0.28)

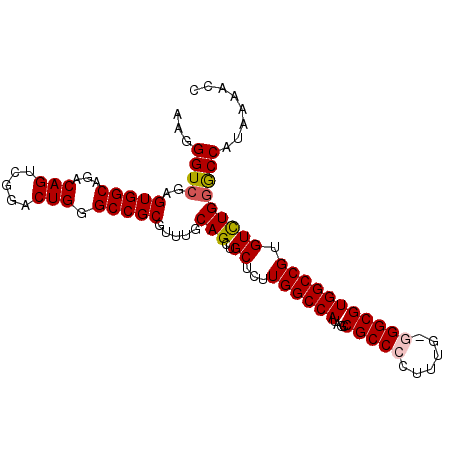
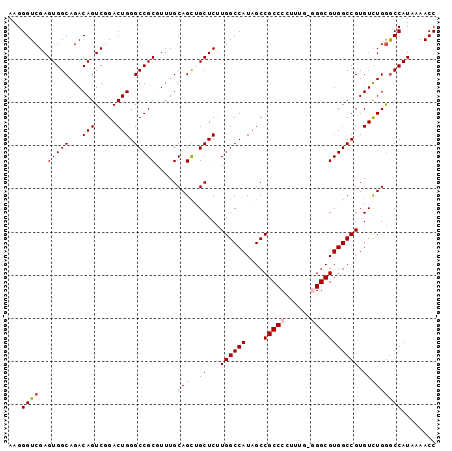
| Location | 17,735,340 – 17,735,439 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 93.63 |
| Mean single sequence MFE | -41.62 |
| Consensus MFE | -35.14 |
| Energy contribution | -36.10 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17735340 99 - 23771897 GGUUUUAUGGCUCAGACACGGCCACGCCCCAAAGGGGGCGGCUAUGGCCAAGAGCAGCUGCAAAUGCGGCCCAGUCCGACUGUCUGCCACUCGACCCUU ((((...((((..(((((.(((((((((((....))))))....)))))..(..(.(((((....)))))...)..)...)))))))))...))))... ( -43.20) >DroSec_CAF1 16462 99 - 1 GGUUUUAUGGCCCAAACACGGCCACGCCGUCAAAGGGGCGGCUAUGGCCAAGAGCCGCUGCAAACGCGGCCCAGUCCGACUGUCUGCCACUCGACCCUU ((((...((((........(((((.((((((.....))))))..))))).((((((((.......))))).(((.....))))))))))...))))... ( -40.20) >DroSim_CAF1 19179 99 - 1 GGUUUUAUGGCCCAGACACGGCCACGCCGCCAAAGGGGCGGCUAUGGCCAAGAGCAGCUGCAAACGCGGCCCAGUCCGACUGUCUGCCACUCGACCCUU ((((...((((..(((((.(((((.((((((.....))))))..)))))..(..(.(((((....)))))...)..)...)))))))))...))))... ( -43.80) >DroEre_CAF1 31388 98 - 1 GGUUUUAUGGUCCAGACACGGCCACGCCC-CAAAUGGGCGGCUAUGGCCAAGAGCCACUGCAAACGCGGCCCAGUCCGACUGUCUGCCACUCGACCCUU ((((...(((..(((((..((((.(((((-.....)))))((..((((.....))))..))......)))).((.....))))))))))...))))... ( -37.90) >DroYak_CAF1 32006 98 - 1 GGUUUUAUGGCCCAGACACGGCCACGCCC-AAAAAGGGCGGCUAUGGCCAGGAGCAGCUGCAAAUGCGGCCCAGUCCGACUGUCUGCCACUCGACCCUU ((((...((((..(((((.((((((((((-.....)))))....))))).(((...(((((....)))))....)))...)))))))))...))))... ( -43.00) >consensus GGUUUUAUGGCCCAGACACGGCCACGCCC_CAAAGGGGCGGCUAUGGCCAAGAGCAGCUGCAAACGCGGCCCAGUCCGACUGUCUGCCACUCGACCCUU ((((...((((..(((((.(((((((((((....))))))....)))))..(..(.(((((....)))))...)..)...)))))))))...))))... (-35.14 = -36.10 + 0.96)

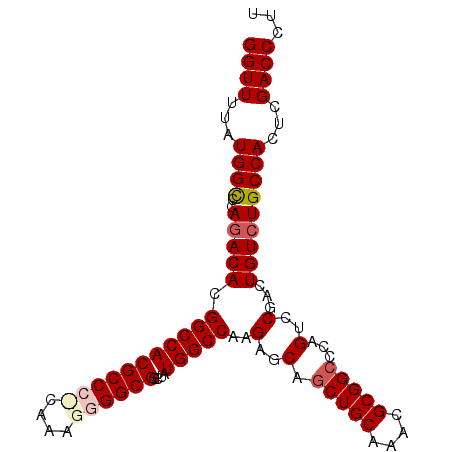
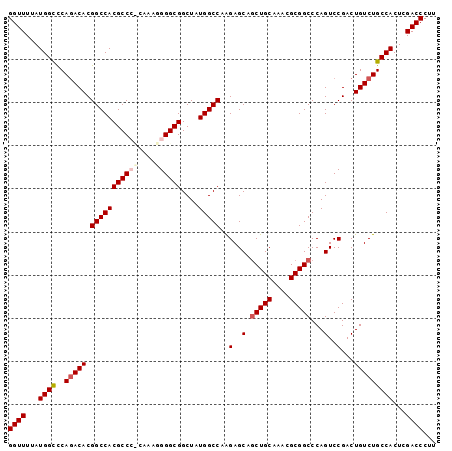
| Location | 17,735,359 – 17,735,479 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -50.06 |
| Consensus MFE | -43.70 |
| Energy contribution | -44.54 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17735359 120 + 23771897 GUCGGACUGGGCCGCAUUUGCAGCUGCUCUUGGCCAUAGCCGCCCCCUUUGGGGCGUGGCCGUGUCUGAGCCAUAAAACCCGCCGCUGGCAAGAGUCAUAAAAAGCCAGUCAAGAGACGG (((.((((((((.((....)).)).(((((((.(((..((.(((((....)))))(((((((....)).))))).......))...)))))))))).........))))))....))).. ( -48.60) >DroSec_CAF1 16481 120 + 1 GUCGGACUGGGCCGCGUUUGCAGCGGCUCUUGGCCAUAGCCGCCCCUUUGACGGCGUGGCCGUGUUUGGGCCAUAAAACCCGCCGCUGGCAAGAGUCAUAAAAAGCCAGUCAAGAGACGG (((.((((((((.((....)).))((((((((.(((..(((((......).))))(((((.(.((((........))))).))))))))))))))))........))))))....))).. ( -51.00) >DroSim_CAF1 19198 120 + 1 GUCGGACUGGGCCGCGUUUGCAGCUGCUCUUGGCCAUAGCCGCCCCUUUGGCGGCGUGGCCGUGUCUGGGCCAUAAAACCCGCCGCUGGCAAGAGUCAUAAAAAGCCAGUCAAGAGACGG (((.((((((((.((....)).)).((((((((((((.((((((.....))))))))))))......(((........)))(((...))))))))).........))))))....))).. ( -53.20) >DroEre_CAF1 31407 119 + 1 GUCGGACUGGGCCGCGUUUGCAGUGGCUCUUGGCCAUAGCCGCCCAUUUG-GGGCGUGGCCGUGUCUGGACCAUAAAACCCGCCGUUGGCAAGAGUCAUAAAAAGCCAGUCAAGAGACGG (((.((((((...((....)).((((((((((.(((..((.((((.....-))))(((((((....))).)))).......))...)))))))))))))......))))))....))).. ( -49.40) >DroYak_CAF1 32025 119 + 1 GUCGGACUGGGCCGCAUUUGCAGCUGCUCCUGGCCAUAGCCGCCCUUUUU-GGGCGUGGCCGUGUCUGGGCCAUAAAACCCGCCGUUGGCAAGAGUCAUAAAAAGCCAGUCAAGAGACGG (((.((((((((.((....)).)).((((...((((..((.((((.....-))))((((((.......)))))).......))...))))..)))).........))))))....))).. ( -48.10) >consensus GUCGGACUGGGCCGCGUUUGCAGCUGCUCUUGGCCAUAGCCGCCCCUUUG_GGGCGUGGCCGUGUCUGGGCCAUAAAACCCGCCGCUGGCAAGAGUCAUAAAAAGCCAGUCAAGAGACGG (((.((((((((.((....)).)).(((((((.(((..((.((((......))))((((((.......)))))).......))...)))))))))).........))))))....))).. (-43.70 = -44.54 + 0.84)

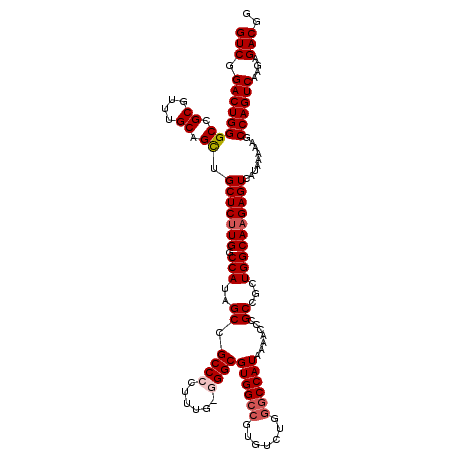
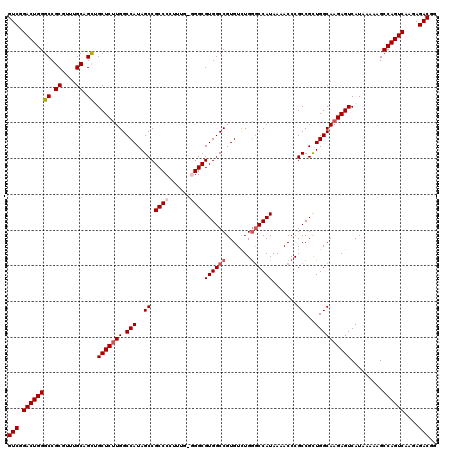
| Location | 17,735,359 – 17,735,479 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -50.79 |
| Consensus MFE | -45.98 |
| Energy contribution | -46.42 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.45 |
| SVM RNA-class probability | 0.999233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17735359 120 - 23771897 CCGUCUCUUGACUGGCUUUUUAUGACUCUUGCCAGCGGCGGGUUUUAUGGCUCAGACACGGCCACGCCCCAAAGGGGGCGGCUAUGGCCAAGAGCAGCUGCAAAUGCGGCCCAGUCCGAC ..(((....((((((.......((.(((((((((..(((........(((((.......)))))((((((....))))))))).))).))))))))(((((....))))))))))).))) ( -53.40) >DroSec_CAF1 16481 120 - 1 CCGUCUCUUGACUGGCUUUUUAUGACUCUUGCCAGCGGCGGGUUUUAUGGCCCAAACACGGCCACGCCGUCAAAGGGGCGGCUAUGGCCAAGAGCCGCUGCAAACGCGGCCCAGUCCGAC ..(((....((((((...............((((((((((((((....)))))......(((((.((((((.....))))))..)))))....))))))((....))))))))))).))) ( -52.36) >DroSim_CAF1 19198 120 - 1 CCGUCUCUUGACUGGCUUUUUAUGACUCUUGCCAGCGGCGGGUUUUAUGGCCCAGACACGGCCACGCCGCCAAAGGGGCGGCUAUGGCCAAGAGCAGCUGCAAACGCGGCCCAGUCCGAC ..(((....((((((.......((.((((((((...)))(((((....)))))......(((((.((((((.....))))))..))))))))))))(((((....))))))))))).))) ( -54.20) >DroEre_CAF1 31407 119 - 1 CCGUCUCUUGACUGGCUUUUUAUGACUCUUGCCAACGGCGGGUUUUAUGGUCCAGACACGGCCACGCCC-CAAAUGGGCGGCUAUGGCCAAGAGCCACUGCAAACGCGGCCCAGUCCGAC ..(((....((((((.......((.(((((((((..(((.(((((........)))).).))).(((((-.....)))))....))).)))))).))((((....)))).)))))).))) ( -43.70) >DroYak_CAF1 32025 119 - 1 CCGUCUCUUGACUGGCUUUUUAUGACUCUUGCCAACGGCGGGUUUUAUGGCCCAGACACGGCCACGCCC-AAAAAGGGCGGCUAUGGCCAGGAGCAGCUGCAAAUGCGGCCCAGUCCGAC ..(((....((((((.......((.(((((((((..(((........(((((.......)))))(((((-.....)))))))).))).))))))))(((((....))))))))))).))) ( -50.30) >consensus CCGUCUCUUGACUGGCUUUUUAUGACUCUUGCCAGCGGCGGGUUUUAUGGCCCAGACACGGCCACGCCC_CAAAGGGGCGGCUAUGGCCAAGAGCAGCUGCAAACGCGGCCCAGUCCGAC ..(((....((((((........(.(((((((((..(((........(((((.......)))))((((((....))))))))).))).))))))).(((((....))))))))))).))) (-45.98 = -46.42 + 0.44)

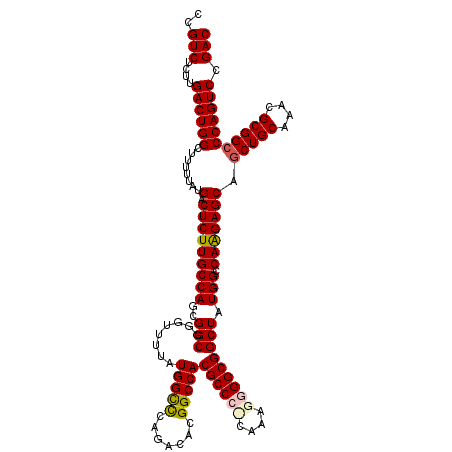
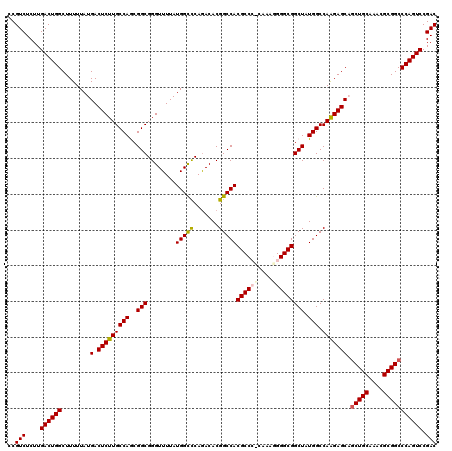
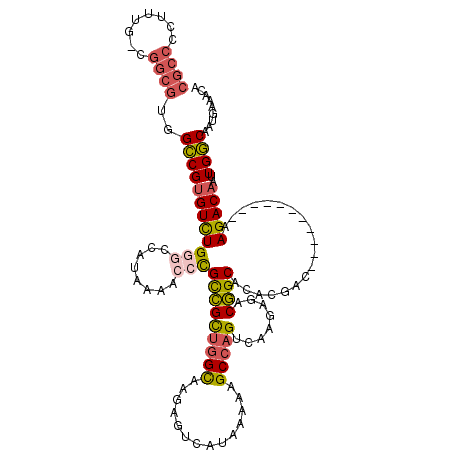
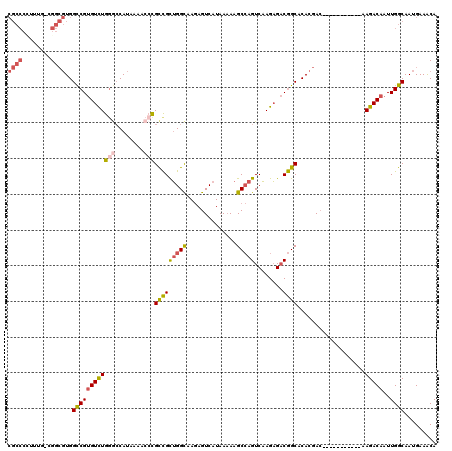
| Location | 17,735,399 – 17,735,508 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.81 |
| Mean single sequence MFE | -33.22 |
| Consensus MFE | -22.68 |
| Energy contribution | -23.19 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17735399 109 + 23771897 CGCCCCCUUUGGGGCGUGGCCGUGUCUGAGCCAUAAAACCCGCCGCUGGCAAGAGUCAUAAAAAGCCAGUCAAGAGACGGCACACGAC-----------AAGACAAUUGGCAAUGAAACA .(((((....)))))((((((((.((((.((..........)))((((((..............))))))..))).))))).)))..(-----------((.....)))........... ( -35.64) >DroVir_CAF1 21774 108 + 1 -----------CGGGCUGGUCGCGUCUGGGGCAAUAAAAGUGUUGCGGGUAAAAGUCAUAA-AAGCAAGUCCAGUGCCAGCACAAGAGAGCACAGAAUGAAGACAAUUGACAAUGAAGCA -----------...(((.((((((.(((((((((((....))))))........((.....-..))...))))))))..((........)).................))).....))). ( -21.00) >DroSec_CAF1 16521 109 + 1 CGCCCCUUUGACGGCGUGGCCGUGUUUGGGCCAUAAAACCCGCCGCUGGCAAGAGUCAUAAAAAGCCAGUCAAGAGACGGCACACGAC-----------AAGACAAUUGGCAAUGAAACA .(((...(((.(..(((((((((.((((((........)))...((((((..............))))))..))).))))).))))..-----------..).)))..)))......... ( -35.24) >DroSim_CAF1 19238 109 + 1 CGCCCCUUUGGCGGCGUGGCCGUGUCUGGGCCAUAAAACCCGCCGCUGGCAAGAGUCAUAAAAAGCCAGUCAAGAGACGGCACACGAC-----------AAGACAAUUGGCAAUGAAACA .(((...(((...((((((((((.((((((........)))...((((((..............))))))..))).))))).)))).)-----------....)))..)))......... ( -38.54) >DroEre_CAF1 31447 108 + 1 CGCCCAUUUG-GGGCGUGGCCGUGUCUGGACCAUAAAACCCGCCGUUGGCAAGAGUCAUAAAAAGCCAGUCAAGAGACGGCACACGAC-----------AAGACAAUUGGCAAUGAAACA .(((...(((-..((((((((((.(((.(((.....(((.....)))(((..............))).))).))).))))).)))).)-----------....)))..)))......... ( -32.44) >DroYak_CAF1 32065 108 + 1 CGCCCUUUUU-GGGCGUGGCCGUGUCUGGGCCAUAAAACCCGCCGUUGGCAAGAGUCAUAAAAAGCCAGUCAAGAGACGGCACACGAC-----------AAGACAAUUGGCAAUGAAACA (((((.....-)))))..(((.((((((((........)))(((..((((..............))))(((....)))))).......-----------.)))))...)))......... ( -36.44) >consensus CGCCCCUUUG_CGGCGUGGCCGUGUCUGGGCCAUAAAACCCGCCGCUGGCAAGAGUCAUAAAAAGCCAGUCAAGAGACGGCACACGAC___________AAGACAAUUGGCAAUGAAACA ((((........))))..((((((((((((........)))(((((((((..............)))))........))))...................)))))..))))......... (-22.68 = -23.19 + 0.51)

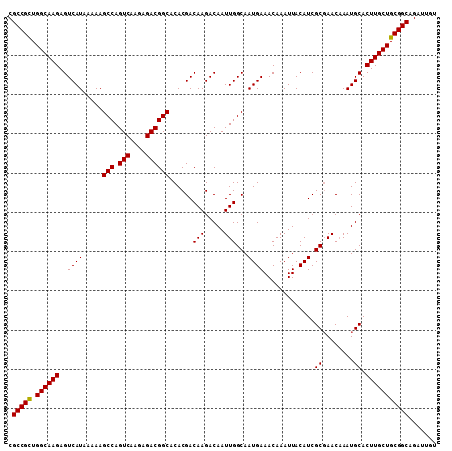
| Location | 17,735,439 – 17,735,548 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 98.35 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -29.98 |
| Energy contribution | -29.74 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.62 |
| SVM RNA-class probability | 0.999463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17735439 109 + 23771897 CGCCGCUGGCAAGAGUCAUAAAAAGCCAGUCAAGAGACGGCACACGACAAGACAAUUGGCAAUGAAACAAAUUACAUCGCUAACAAAUGCACUUGCUGCGGCAGAUUGU .(((((.((((((.(.(((.....(((.(((....))))))..............(((((.(((..........))).)))))...)))).)))))))))))....... ( -33.10) >DroSec_CAF1 16561 109 + 1 CGCCGCUGGCAAGAGUCAUAAAAAGCCAGUCAAGAGACGGCACACGACAAGACAAUUGGCAAUGAAACAAAUUACAUCGCGAACAAAUGCACUUGCUGCGGCAGAUUGU .(((((.((((((.(.(((.....(((.(((....))))))..............((.((.(((..........))).)).))...)))).)))))))))))....... ( -31.00) >DroSim_CAF1 19278 109 + 1 CGCCGCUGGCAAGAGUCAUAAAAAGCCAGUCAAGAGACGGCACACGACAAGACAAUUGGCAAUGAAACAAAUUACAUCGCGAACAAAUGCACUUGCUGCGGCAGACUGU .(((((.((((((.(.(((.....(((.(((....))))))..............((.((.(((..........))).)).))...)))).)))))))))))....... ( -31.00) >DroEre_CAF1 31486 109 + 1 CGCCGUUGGCAAGAGUCAUAAAAAGCCAGUCAAGAGACGGCACACGACAAGACAAUUGGCAAUGAAACAAAUUACAUCGCGAACCAAUGCACUUGCUGCGGCAGAUUGU .(((((.((((((.(.(((.....(((.(((....))))))..............((.((.(((..........))).)).))...)))).)))))))))))....... ( -28.40) >DroYak_CAF1 32104 109 + 1 CGCCGUUGGCAAGAGUCAUAAAAAGCCAGUCAAGAGACGGCACACGACAAGACAAUUGGCAAUGAAACAAAUUACAUCGCGAACAAAUGCACUUGCUGCGGCAGAUUGU .(((((.((((((.(.(((.....(((.(((....))))))..............((.((.(((..........))).)).))...)))).)))))))))))....... ( -28.40) >consensus CGCCGCUGGCAAGAGUCAUAAAAAGCCAGUCAAGAGACGGCACACGACAAGACAAUUGGCAAUGAAACAAAUUACAUCGCGAACAAAUGCACUUGCUGCGGCAGAUUGU .(((((.((((((..((((.....(((.(((....))))))......(((.....)))...)))).............((........)).)))))))))))....... (-29.98 = -29.74 + -0.24)


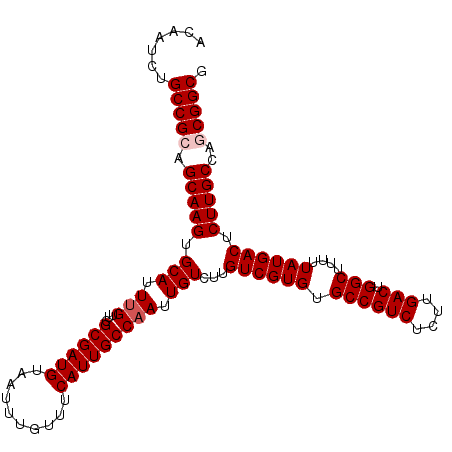
| Location | 17,735,439 – 17,735,548 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 98.35 |
| Mean single sequence MFE | -35.68 |
| Consensus MFE | -33.38 |
| Energy contribution | -33.98 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.70 |
| SVM RNA-class probability | 0.999538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17735439 109 - 23771897 ACAAUCUGCCGCAGCAAGUGCAUUUGUUAGCGAUGUAAUUUGUUUCAUUGCCAAUUGUCUUGUCGUGUGCCGUCUCUUGACUGGCUUUUUAUGACUCUUGCCAGCGGCG .......(((((.(((((.(((.(((...((((((..........))))))))).)))...((((((.((((((....))).)))....)))))).)))))..))))). ( -37.30) >DroSec_CAF1 16561 109 - 1 ACAAUCUGCCGCAGCAAGUGCAUUUGUUCGCGAUGUAAUUUGUUUCAUUGCCAAUUGUCUUGUCGUGUGCCGUCUCUUGACUGGCUUUUUAUGACUCUUGCCAGCGGCG .......(((((.(((((.(((.(((...((((((..........))))))))).)))...((((((.((((((....))).)))....)))))).)))))..))))). ( -37.30) >DroSim_CAF1 19278 109 - 1 ACAGUCUGCCGCAGCAAGUGCAUUUGUUCGCGAUGUAAUUUGUUUCAUUGCCAAUUGUCUUGUCGUGUGCCGUCUCUUGACUGGCUUUUUAUGACUCUUGCCAGCGGCG .......(((((.(((((.(((.(((...((((((..........))))))))).)))...((((((.((((((....))).)))....)))))).)))))..))))). ( -37.30) >DroEre_CAF1 31486 109 - 1 ACAAUCUGCCGCAGCAAGUGCAUUGGUUCGCGAUGUAAUUUGUUUCAUUGCCAAUUGUCUUGUCGUGUGCCGUCUCUUGACUGGCUUUUUAUGACUCUUGCCAACGGCG .......((((..(((((......((((.((((((..........)))))).)))).....((((((.((((((....))).)))....)))))).)))))...)))). ( -32.80) >DroYak_CAF1 32104 109 - 1 ACAAUCUGCCGCAGCAAGUGCAUUUGUUCGCGAUGUAAUUUGUUUCAUUGCCAAUUGUCUUGUCGUGUGCCGUCUCUUGACUGGCUUUUUAUGACUCUUGCCAACGGCG .......((((..(((((.(((.(((...((((((..........))))))))).)))...((((((.((((((....))).)))....)))))).)))))...)))). ( -33.70) >consensus ACAAUCUGCCGCAGCAAGUGCAUUUGUUCGCGAUGUAAUUUGUUUCAUUGCCAAUUGUCUUGUCGUGUGCCGUCUCUUGACUGGCUUUUUAUGACUCUUGCCAGCGGCG .......(((((.(((((.(((.(((...((((((..........))))))))).)))...((((((.((((((....))).)))....)))))).)))))..))))). (-33.38 = -33.98 + 0.60)

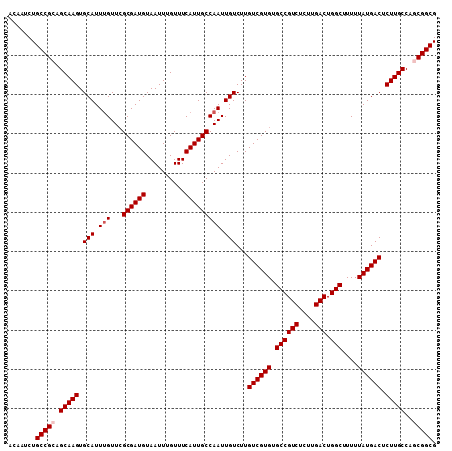
| Location | 17,735,508 – 17,735,608 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 98.20 |
| Mean single sequence MFE | -28.88 |
| Consensus MFE | -27.60 |
| Energy contribution | -28.20 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.993918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17735508 100 - 23771897 CAUUGCGAAUUCAGCAACGAGCAAGAACACGAAAUGUUCUUCCAUUUAGUUGCGCCAAGGACAAUCUGCCGCAGCAAGUGCAUUUGUUAGCGAUGUAAUU ((((((.(((...(((....(.(((((((.....))))))).).....((((((...((......))..))))))...)))....))).))))))..... ( -24.60) >DroSec_CAF1 16630 100 - 1 CAUUGCGAAUUCAGCAACGAGCAAGAACACGAAAUGUUCUUCCAUUUAGUUGCGCCAAGGACAAUCUGCCGCAGCAAGUGCAUUUGUUCGCGAUGUAAUU ((((((((((...(((....(.(((((((.....))))))).).....((((((...((......))..))))))...)))....))))))))))..... ( -30.50) >DroSim_CAF1 19347 100 - 1 CAUUGCGAAUUCAGCAACGAGCAAGAACACGAAAUGUUCUUCCAUUUAGUUGCGCCAAGGACAGUCUGCCGCAGCAAGUGCAUUUGUUCGCGAUGUAAUU ((((((((((...(((....(.(((((((.....))))))).).....((((((...(((....)))..))))))...)))....))))))))))..... ( -30.90) >DroEre_CAF1 31555 100 - 1 CAUUGCGAAUUCAGCAACGAGCAAGAACACGAAAUGUUCCUCCAUUUAGUUGCGCCAAGGACAAUCUGCCGCAGCAAGUGCAUUGGUUCGCGAUGUAAUU (((((((((((..(((..(((...(((((.....))))))))......((((((...((......))..))))))...)))...)))))))))))..... ( -29.30) >DroYak_CAF1 32173 100 - 1 CAUUGCGAAUUCAGCAACGAGCAAGAACACGAAAUGUUCCUCCAUUUAGUUGCGCCAAGGACAAUCUGCCGCAGCAAGUGCAUUUGUUCGCGAUGUAAUU ((((((((((...(((..(((...(((((.....))))))))......((((((...((......))..))))))...)))....))))))))))..... ( -29.10) >consensus CAUUGCGAAUUCAGCAACGAGCAAGAACACGAAAUGUUCUUCCAUUUAGUUGCGCCAAGGACAAUCUGCCGCAGCAAGUGCAUUUGUUCGCGAUGUAAUU ((((((((((...(((....(.(((((((.....))))))).).....((((((...((......))..))))))...)))....))))))))))..... (-27.60 = -28.20 + 0.60)

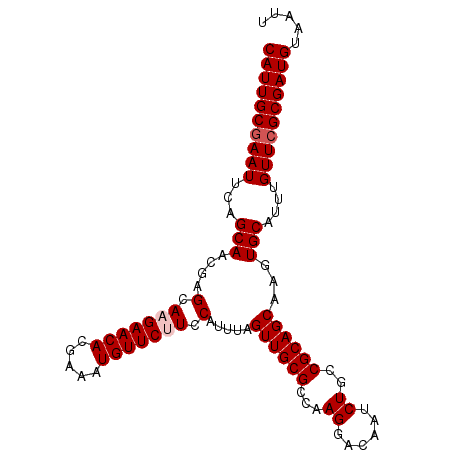
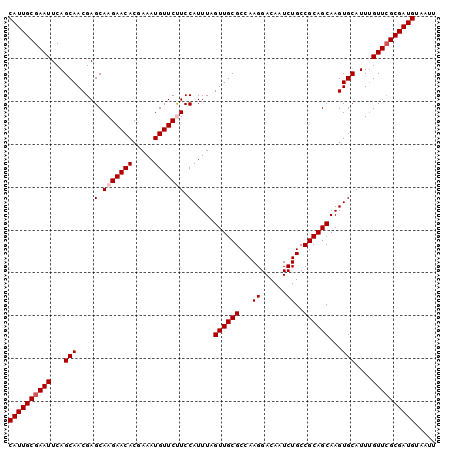
| Location | 17,735,608 – 17,735,710 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -22.96 |
| Consensus MFE | -15.92 |
| Energy contribution | -15.76 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17735608 102 - 23771897 UAAAAAGCACUUAUAUCCUGCAAUGAAAUCAUUAUCAAGAGUAACCUGUCUUUGUUUCUCCAGUGUAAUUGCAUGAAAAUUGUGACAUCACAUUGGAGCCAU ...((((((......((.((.((((....))))..)).))......)).))))....(((((((((...((((..(...)..)).))..))))))))).... ( -17.90) >DroSec_CAF1 16730 102 - 1 CAAAAAGCAGUUUUAUCCAGCAAUGAAAUCAAUUUCAAGAGUAACCUGUCUUGGUUUCUCCAGUGUAAUUGCAUGAAAAUUGUGACAUCACAUUGGAGCCAU ...............(((((((((((((....))))..(((.((((......)))).))).......)))))........((((....)))).))))..... ( -20.80) >DroSim_CAF1 19447 102 - 1 CAAAAAGCAGUUUUAUCCAGCAAUGAAAUCAAUUUCAAGAGUAACCUGUCUUGGUUUCUCCAGUGUAAUUGCAUGACAAUUGUGACAUCACAUUGGAGCCAU ...............(((((((((((((....))))..(((.((((......)))).))).......)))))........((((....)))).))))..... ( -20.80) >DroEre_CAF1 31655 101 - 1 UACAAAGUUGUUUUGGCCAGCCCUGAAACUAUUUUCAGGAGUAACCU-GUCUUGUUUCUCCAGUGUAAGUGCACGGGAAUUGUGACAUCACAUUGGAGCCAU .............((((..(((((((((....))))))).)).....-..........((((((((..(((((((.....)))).))).)))))))))))). ( -28.20) >DroYak_CAF1 32273 99 - 1 U---AAGUCUUUUUGACAAGAAACGAAAUCAUUUUCAAGAGUAUCCUCGCCUUCUUCCUCCAGUGUAAUUGCACGAAAAUUGUGACAUCACACUGGAGCCAU .---..(((.....)))(((((..((((....))))..(((....)))...))))).(((((((((...(((((((...))))).))..))))))))).... ( -27.10) >consensus UAAAAAGCAGUUUUAUCCAGCAAUGAAAUCAUUUUCAAGAGUAACCUGUCUUUGUUUCUCCAGUGUAAUUGCAUGAAAAUUGUGACAUCACAUUGGAGCCAU .......................(((((....)))))....................(((((((((...(((((((...))))).))..))))))))).... (-15.92 = -15.76 + -0.16)

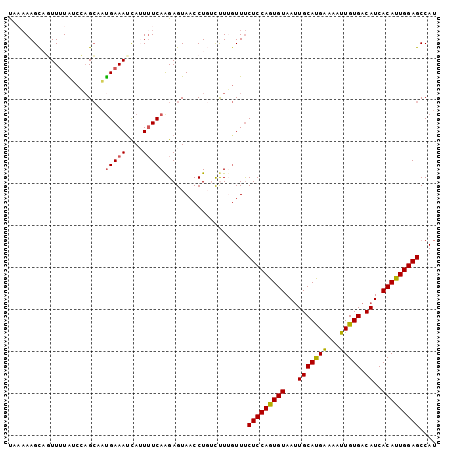
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:57 2006