| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
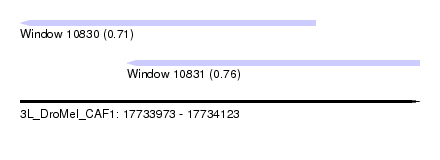
| Location | 17,733,973 – 17,734,123 |
| Length | 150 |
| Max. P | 0.758742 |

| Location | 17,733,973 – 17,734,084 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 91.10 |
| Mean single sequence MFE | -32.07 |
| Consensus MFE | -24.93 |
| Energy contribution | -24.99 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
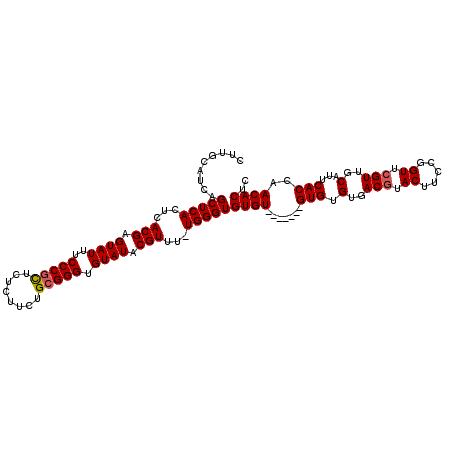
>3L_DroMel_CAF1 17733973 111 - 23771897 UGCGGGUGUAUACGUUU-UGGGUGUGU--G-UGUGUGUGUGACGUACUUCCGGUUCGUUGCAUUCACCAACACUCGCGUACACCUCCUUUACAUGCAUUCUAUUUCAGAUCUACC (((((((((..((((..-.((((((.(--(-.(((.(((..(((.((.....)).)))..))).))))))))))))))))))))).........))).................. ( -33.10) >DroSec_CAF1 15116 107 - 1 UGCGGGUGUAUACGUUU-UGGGUGUGU-------GUGUGUGACGUACUUCCGGUUCGUUGCUUUCACCAACACUCGCGCACACCUCCUUUACAUGCAUUCUAUUUCAGAUAUACU ...((((((((......-.((((((((-------(((.(..(((.((.....)).)))..).............))))))))))).......))))))))............... ( -30.48) >DroSim_CAF1 17817 107 - 1 UGCGGGUGUAUACGUUU-UGGGUGUGU-------GUGUGUGACGUACUUCCGGUUAGUUGCUUUCACCAACACUCGUGCACACCUCCUUUACAUGCAUUCUAUUUCAGAUAUACU ...((((((((......-.(((((((.-------.((.(((..(......)(((.((.....)).)))..))).))..))))))).......))))))))............... ( -27.34) >DroYak_CAF1 30595 115 - 1 CGCGGGUGUAUACGUUUAUGGGUGUGUGUGUUGUGUGUGUGACGUACUUCCGGUUCGUUGCAUUCACCAACACUUGCGCACACCUCCUCUUCAUGCAUUCUAUUUCAGAUAUACC ...((((((((........((((((((((((((((.(((..(((.((.....)).)))..))).)).))))))....)))))))).......))))))))............... ( -37.36) >consensus UGCGGGUGUAUACGUUU_UGGGUGUGU_______GUGUGUGACGUACUUCCGGUUCGUUGCAUUCACCAACACUCGCGCACACCUCCUUUACAUGCAUUCUAUUUCAGAUAUACC ...((((((((........((((((((.......(((.(..(((.((.....)).)))..)...)))..........)))))))).......))))))))............... (-24.93 = -24.99 + 0.06)



| Location | 17,734,013 – 17,734,123 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 91.47 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -27.92 |
| Energy contribution | -28.18 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.758742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17734013 110 - 23771897 CUCGCAUCAGCUCACUCACGAGUAUUUCCCGCUCUCUUCUGCGGGUGUAUACGUUU-UGGGUGUGU--G-UGUGUGUGUGACGUACUUCCGGUUCGUUGCAUUCACCAACACUC ...((....)).((((((.((((((..(((((........)))))...)))).)).-)))))).((--(-((((.(((..(((.((.....)).)))..))).)))..)))).. ( -38.10) >DroSec_CAF1 15156 106 - 1 CUUGCAUCAGCUCACUCACGAGUAUUUCCCGUUCUCUUCUGCGGGUGUAUACGUUU-UGGGUGUGU-------GUGUGUGACGUACUUCCGGUUCGUUGCUUUCACCAACACUC .........((.((((((.((((((..(((((........)))))...)))).)).-)))))).))-------(((.(..(((.((.....)).)))..)...)))........ ( -30.50) >DroSim_CAF1 17857 106 - 1 CUUGCAUCAGCUCACUCACGAGUAUUUCCCGUUCUCUUCUGCGGGUGUAUACGUUU-UGGGUGUGU-------GUGUGUGACGUACUUCCGGUUAGUUGCUUUCACCAACACUC .........((.((((((.((((((..(((((........)))))...)))).)).-)))))).))-------((((((((.(((((.......)).)))..))))..)))).. ( -27.90) >DroYak_CAF1 30635 114 - 1 CCUGCAUCAGCUCACUCACGAGUAUUUCCCCCUCCCUUCCGCGGGUGUAUACGUUUAUGGGUGUGUGUGUUGUGUGUGUGACGUACUUCCGGUUCGUUGCAUUCACCAACACUU .........((.((((((...((((..(((.(........).)))...)))).....)))))).))((((((((.(((..(((.((.....)).)))..))).)).)))))).. ( -34.70) >consensus CUUGCAUCAGCUCACUCACGAGUAUUUCCCGCUCUCUUCUGCGGGUGUAUACGUUU_UGGGUGUGU_______GUGUGUGACGUACUUCCGGUUCGUUGCAUUCACCAACACUC .........(((((...(((.((((..(((((........))))).)))).)))...)))))((((.......(((.(..(((.((.....)).)))..)...)))..)))).. (-27.92 = -28.18 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:47 2006