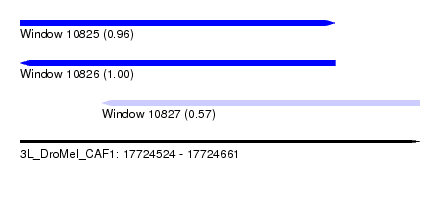
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,724,524 – 17,724,661 |
| Length | 137 |
| Max. P | 0.999834 |

| Location | 17,724,524 – 17,724,632 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -36.64 |
| Consensus MFE | -30.82 |
| Energy contribution | -30.90 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
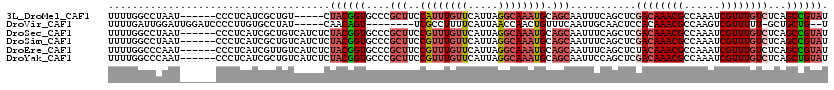
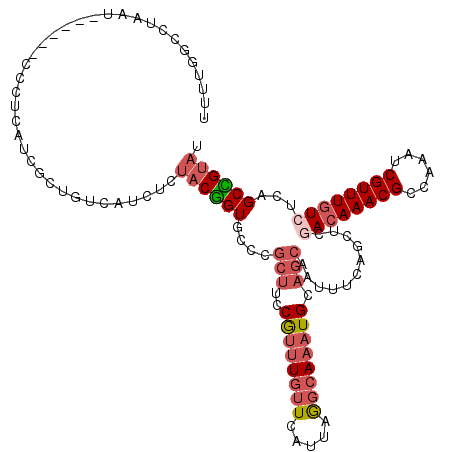
>3L_DroMel_CAF1 17724524 108 + 23771897 GUAGAAGCGGAUAGGGGUGCGG------------AUGCGGAUACGGCUGAGACAAACGAUUUGGCGUUUGUCGAGCUGAAAUUGCUGCAUUUGCCUAAUGAACAAAUGGAAGCGGGCACC ......(((..((..((.((((------------((((((...(((((..((((((((......)))))))).)))))......))))))))))))..))..)...((....)).))... ( -36.00) >DroSec_CAF1 5853 99 + 1 ---------GAUAGGGGUGCGG------------AUGCGAAUACGGCUGAGACAAACGAUUUGGCGUUUGUCGAGCUGAAAUUGCUGCAUUUGCCUAAUGAACAAACGGAAGCGGGCACC ---------......(((((((------------(((((....(((((..((((((((......)))))))).))))).......)))))))..............((....)).))))) ( -31.50) >DroSim_CAF1 6072 108 + 1 GUAGAAGCGGAUAGGGGUGCGG------------AUGCGGAUACGGCUGAGACAAACGAUUUGGCGUUUGUCGAGCUGAAAUUGCUGCAUUUGCCUAAUGAACAAACGGAAGCGGGCACC ......(((..((..((.((((------------((((((...(((((..((((((((......)))))))).)))))......))))))))))))..))..)...((....)).))... ( -38.20) >DroEre_CAF1 20164 120 + 1 GUAAAAGCGGAUAGGGGUGCGGAAGCGUAUGCGGAUGCGGAUACGGCUGAGACAAACGAUUUGGCGUUUGUAGAGCUGAAAUUGCUGCAUUUGCCUAAUGAACAAACGGAAGCGGGCACC ......((...(((..(((((....)))))((((((((((...(((((...(((((((......)))))))..)))))......))))))))))))).........((....)).))... ( -37.80) >DroYak_CAF1 20552 120 + 1 GUAAAAGCGAAUAGGGGUGCGGAUGCGAAUACGGAUGCGGAUACAGCUGAGACAAACGAUUUGGCGUUUGUCGAGCUGGAAUUGCUGCAUUUGCCUAAUGAACAAACGGAAGCGGGCACC ...............(((((...(((.....(((((((((...(((((..((((((((......)))))))).)))))......)))))))))((............))..))).))))) ( -39.70) >consensus GUAAAAGCGGAUAGGGGUGCGG____________AUGCGGAUACGGCUGAGACAAACGAUUUGGCGUUUGUCGAGCUGAAAUUGCUGCAUUUGCCUAAUGAACAAACGGAAGCGGGCACC ...............(((((..............((((((...(((((..((((((((......)))))))).)))))......))))))................((....)).))))) (-30.82 = -30.90 + 0.08)



| Location | 17,724,524 – 17,724,632 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -30.36 |
| Consensus MFE | -28.48 |
| Energy contribution | -28.52 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.20 |
| SVM RNA-class probability | 0.999834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17724524 108 - 23771897 GGUGCCCGCUUCCAUUUGUUCAUUAGGCAAAUGCAGCAAUUUCAGCUCGACAAACGCCAAAUCGUUUGUCUCAGCCGUAUCCGCAU------------CCGCACCCCUAUCCGCUUCUAC (((((..(((..((((((((.....)))))))).))).......(((.((((((((......))))))))..)))...........------------..)))))............... ( -29.80) >DroSec_CAF1 5853 99 - 1 GGUGCCCGCUUCCGUUUGUUCAUUAGGCAAAUGCAGCAAUUUCAGCUCGACAAACGCCAAAUCGUUUGUCUCAGCCGUAUUCGCAU------------CCGCACCCCUAUC--------- (((((..(((..((((((((.....)))))))).))).......(((.((((((((......))))))))..)))...........------------..)))))......--------- ( -29.40) >DroSim_CAF1 6072 108 - 1 GGUGCCCGCUUCCGUUUGUUCAUUAGGCAAAUGCAGCAAUUUCAGCUCGACAAACGCCAAAUCGUUUGUCUCAGCCGUAUCCGCAU------------CCGCACCCCUAUCCGCUUCUAC (((((..(((..((((((((.....)))))))).))).......(((.((((((((......))))))))..)))...........------------..)))))............... ( -29.40) >DroEre_CAF1 20164 120 - 1 GGUGCCCGCUUCCGUUUGUUCAUUAGGCAAAUGCAGCAAUUUCAGCUCUACAAACGCCAAAUCGUUUGUCUCAGCCGUAUCCGCAUCCGCAUACGCUUCCGCACCCCUAUCCGCUUUUAC (((((..(((..((((((((.....)))))))).))).......(((..(((((((......)))))))...)))(((((.((....)).))))).....)))))............... ( -29.70) >DroYak_CAF1 20552 120 - 1 GGUGCCCGCUUCCGUUUGUUCAUUAGGCAAAUGCAGCAAUUCCAGCUCGACAAACGCCAAAUCGUUUGUCUCAGCUGUAUCCGCAUCCGUAUUCGCAUCCGCACCCCUAUUCGCUUUUAC (((((..(((..((((((((.....)))))))).))).....(((((.((((((((......))))))))..))))).......................)))))............... ( -33.50) >consensus GGUGCCCGCUUCCGUUUGUUCAUUAGGCAAAUGCAGCAAUUUCAGCUCGACAAACGCCAAAUCGUUUGUCUCAGCCGUAUCCGCAU____________CCGCACCCCUAUCCGCUUCUAC (((((..(((..((((((((.....)))))))).))).......(((.((((((((......))))))))..))).........................)))))............... (-28.48 = -28.52 + 0.04)

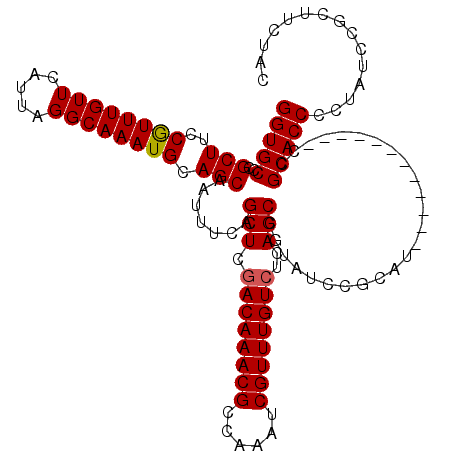
| Location | 17,724,552 – 17,724,661 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.59 |
| Mean single sequence MFE | -26.04 |
| Consensus MFE | -18.49 |
| Energy contribution | -19.85 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17724552 109 - 23771897 UUUUGGCCUAAU------CCCUCAUCGCUGU-----CUACGGUGCCCGCUUCCAUUUGUUCAUUAGGCAAAUGCAGCAAUUUCAGCUCGACAAACGCCAAAUCGUUUGUCUCAGCCGUAU ....(((.....------........((((.-----....((...))(((..((((((((.....)))))))).))).....))))..((((((((......))))))))...))).... ( -28.80) >DroVir_CAF1 6682 104 - 1 UUUUGAUUGGAUUGGAUCCCCUUGUGCCUAU-----CAACAGU--------UCGCCUUUUCAUUAACCAACUGUUUCAAUUGCAACUCCACAAACGCCAAGUCGUUUUU-GCUGCUG--U ...((((.((...((....)).....)).))-----))(((((--------(.(............).)))))).......(((.(..((.(((((......))))).)-)..).))--) ( -13.50) >DroSec_CAF1 5872 114 - 1 UUUUGGCCUAAU------CCCUCAUCGCUGUCAUCUCUACGGUGCCCGCUUCCGUUUGUUCAUUAGGCAAAUGCAGCAAUUUCAGCUCGACAAACGCCAAAUCGUUUGUCUCAGCCGUAU ....(((.....------........((((.((((.....))))...(((..((((((((.....)))))))).))).....))))..((((((((......))))))))...))).... ( -30.10) >DroSim_CAF1 6100 114 - 1 UUUUGGCCUAAU------CCCUCAUCGCUGUCAUCUCUACGGUGCCCGCUUCCGUUUGUUCAUUAGGCAAAUGCAGCAAUUUCAGCUCGACAAACGCCAAAUCGUUUGUCUCAGCCGUAU ....(((.....------........((((.((((.....))))...(((..((((((((.....)))))))).))).....))))..((((((((......))))))))...))).... ( -30.10) >DroEre_CAF1 20204 114 - 1 UUUUGGCCCAAU------CCCUCAUCGUUGUCAUCUCUACGGUGCCCGCUUCCGUUUGUUCAUUAGGCAAAUGCAGCAAUUUCAGCUCUACAAACGCCAAAUCGUUUGUCUCAGCCGUAU ....(((.....------....((((((.(......).))))))...(((..((((((((.....)))))))).)))............(((((((......)))))))....))).... ( -24.20) >DroYak_CAF1 20592 114 - 1 UUUUGGCCCAAU------CCCUCAUCGCUGUCAUCUCUACGGUGCCCGCUUCCGUUUGUUCAUUAGGCAAAUGCAGCAAUUCCAGCUCGACAAACGCCAAAUCGUUUGUCUCAGCUGUAU ....(((((...------......................)).))).(((..((((((((.....)))))))).))).....(((((.((((((((......))))))))..)))))... ( -29.51) >consensus UUUUGGCCUAAU______CCCUCAUCGCUGUCAUCUCUACGGUGCCCGCUUCCGUUUGUUCAUUAGGCAAAUGCAGCAAUUUCAGCUCGACAAACGCCAAAUCGUUUGUCUCAGCCGUAU .....................................((((((....(((..((((((((.....)))))))).)))...........((((((((......))))))))...)))))). (-18.49 = -19.85 + 1.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:43 2006