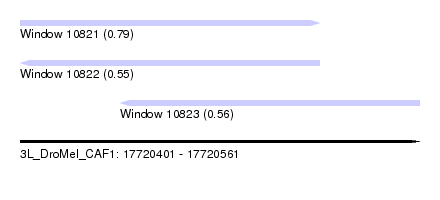
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,720,401 – 17,720,561 |
| Length | 160 |
| Max. P | 0.785442 |

| Location | 17,720,401 – 17,720,521 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.06 |
| Mean single sequence MFE | -41.80 |
| Consensus MFE | -30.61 |
| Energy contribution | -31.58 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
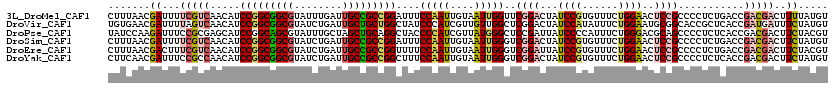
>3L_DroMel_CAF1 17720401 120 + 23771897 ACAUAAAAGUCGUCGGUCAGAGGGGCGGAGUUCCAGAAACACGGAUAGUCCGAACCAAUUACAAUUGGAAAUCCGGCGGCAAUCAAAUACGCCGCCGGAUGUUGACGAAAAUCGUUAAAG .........(((((((.(...((..((((.((((........))).).))))..))..............((((((((((..........))))))))))))))))))............ ( -42.40) >DroVir_CAF1 1971 120 + 1 ACAUAGAAAUCAUCGGUGAGCGGUGCCGCAUUCCAGAAAUAUGGAUAGUCCGAGCCAACAACGAUGGGAUAGCCAGCAGCAAUCAGAUACGCCGCCGGAUGUUGACUAAAAUCGUUCACA ...............(((((((((.......((((......))))(((((....(((.......)))((((.((.((.((..........)).)).)).)))))))))..))))))))). ( -30.10) >DroPse_CAF1 4127 120 + 1 ACGUAGAAGUCGUCGGUGAGAGGGGCUGCGUCCCAGAAAUGGGGAUAAUCGGAGCCCAUAACGAUGGGGUAGCCUGCAGCUAGCAAAUACGCUGCCGGAUGCUCGCGGAAAUCUUGGAUA ..((..(((...((.(((((..(((((((.(((((....)))))..........(((((....))))))))))))(((((((.....)).)))))......))))).))...)))..)). ( -45.30) >DroSim_CAF1 1667 120 + 1 ACAUAGAAGUCGUCGGUCAGAGGGGCGGAGUUCCAGAAACACGGAUAGUCCGACCCAAUUACAAUUGGAAAUCCGGCGGCAAUCAGAUACGCCGCCGGAUGUUGACGAAAAUCGUUAAAG .....((..(((((((.(...(((.((((.((((........))).).)))).)))..............((((((((((..........))))))))))))))))))...))....... ( -44.60) >DroEre_CAF1 16044 120 + 1 ACGUAGAAGUCGUCGGUCAGAGGGGCGGAGUUCCAGAAACACGGAUAAUCCGACCCAAUUACAAUUGGAAAACCGGCGGCAAUCAGAUACGCCGCCGGAUGUUGACGAAAGUCGUUAAAG .....((..(((((((.((..(((.((((..(((........)))...)))).)))................((((((((..........)))))))).)))))))))...))....... ( -44.90) >DroYak_CAF1 16350 120 + 1 ACAUAGAAGUCGUCGGUGAGAGGGGCGGAGUUCCAGAAACACGGAUAGUCCGACCCAAUUACAAUUGGAAAGCCGGCGGCAAUCAGAUACGCCGCCGGAUGUUGGCGGAAAUCGUUGAAG .....((..((((((((((..(((.((((.((((........))).).)))).)))..))))..........((((((((..........))))))))....))))))...))....... ( -43.50) >consensus ACAUAGAAGUCGUCGGUCAGAGGGGCGGAGUUCCAGAAACACGGAUAGUCCGACCCAAUUACAAUUGGAAAGCCGGCGGCAAUCAGAUACGCCGCCGGAUGUUGACGAAAAUCGUUAAAG .....((..(((((((.....(((.((((..(((........)))...)))).)))................((((((((..........))))))))...)))))))...))....... (-30.61 = -31.58 + 0.97)



| Location | 17,720,401 – 17,720,521 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.06 |
| Mean single sequence MFE | -37.59 |
| Consensus MFE | -25.70 |
| Energy contribution | -26.68 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17720401 120 - 23771897 CUUUAACGAUUUUCGUCAACAUCCGGCGGCGUAUUUGAUUGCCGCCGGAUUUCCAAUUGUAAUUGGUUCGGACUAUCCGUGUUUCUGGAACUCCGCCCCUCUGACCGACGACUUUUAUGU ............(((((...(((((((((((........)))))))))))........((....((..((((...((((......))))..))))..))....)).)))))......... ( -40.30) >DroVir_CAF1 1971 120 - 1 UGUGAACGAUUUUAGUCAACAUCCGGCGGCGUAUCUGAUUGCUGCUGGCUAUCCCAUCGUUGUUGGCUCGGACUAUCCAUAUUUCUGGAAUGCGGCACCGCUCACCGAUGAUUUCUAUGU .......((..((((((((((.(((((((((........)))))))))............)))))))((((....((((......))))..(((....)))...)))))))..))..... ( -35.42) >DroPse_CAF1 4127 120 - 1 UAUCCAAGAUUUCCGCGAGCAUCCGGCAGCGUAUUUGCUAGCUGCAGGCUACCCCAUCGUUAUGGGCUCCGAUUAUCCCCAUUUCUGGGACGCAGCCCCUCUCACCGACGACUUCUACGU ......(((..((..(((((...((((((((....)))).))))...))).............(((((.((....((((.......)))))).))))).......))..))..))).... ( -31.30) >DroSim_CAF1 1667 120 - 1 CUUUAACGAUUUUCGUCAACAUCCGGCGGCGUAUCUGAUUGCCGCCGGAUUUCCAAUUGUAAUUGGGUCGGACUAUCCGUGUUUCUGGAACUCCGCCCCUCUGACCGACGACUUCUAUGU .......((...(((((...(((((((((((........)))))))))))........((....(((.((((...((((......))))..)))).)))....)).)))))..))..... ( -43.90) >DroEre_CAF1 16044 120 - 1 CUUUAACGACUUUCGUCAACAUCCGGCGGCGUAUCUGAUUGCCGCCGGUUUUCCAAUUGUAAUUGGGUCGGAUUAUCCGUGUUUCUGGAACUCCGCCCCUCUGACCGACGACUUCUACGU ............(((((.....(((((((((........)))))))))..........((....(((.((((...((((......))))..)))).)))....)).)))))......... ( -40.50) >DroYak_CAF1 16350 120 - 1 CUUCAACGAUUUCCGCCAACAUCCGGCGGCGUAUCUGAUUGCCGCCGGCUUUCCAAUUGUAAUUGGGUCGGACUAUCCGUGUUUCUGGAACUCCGCCCCUCUCACCGACGACUUCUAUGU .......((..((.........(((((((((........)))))))))................(((.((((...((((......))))..)))).)))..........))..))..... ( -34.10) >consensus CUUUAACGAUUUUCGUCAACAUCCGGCGGCGUAUCUGAUUGCCGCCGGCUUUCCAAUUGUAAUUGGGUCGGACUAUCCGUGUUUCUGGAACUCCGCCCCUCUCACCGACGACUUCUAUGU .......((...(((((.....(((((((((........)))))))))....(((((....)))))..((((...((((......))))..))))...........)))))..))..... (-25.70 = -26.68 + 0.98)
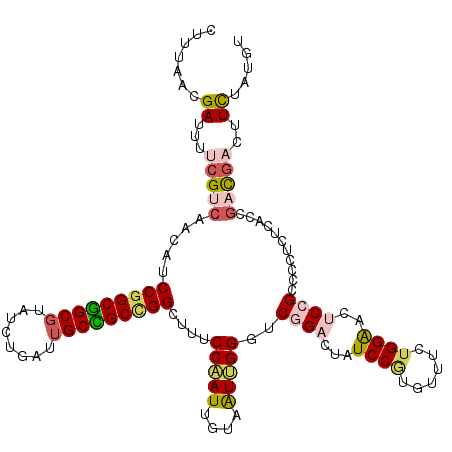
| Location | 17,720,441 – 17,720,561 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.89 |
| Mean single sequence MFE | -36.38 |
| Consensus MFE | -26.33 |
| Energy contribution | -26.28 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17720441 120 - 23771897 CCGUGGAGAUCUGUCCGCUGUCCAACCAUUAUCUGCAAUACUUUAACGAUUUUCGUCAACAUCCGGCGGCGUAUUUGAUUGCCGCCGGAUUUCCAAUUGUAAUUGGUUCGGACUAUCCGU ..(((((......))))).(((((((((.....((((((......(((.....)))....(((((((((((........))))))))))).....))))))..))))).))))....... ( -43.70) >DroVir_CAF1 2011 120 - 1 CCGCCGAGGUGUGUCCACUGUCCAAUUAUUAUCUGCAGUAUGUGAACGAUUUUAGUCAACAUCCGGCGGCGUAUCUGAUUGCUGCUGGCUAUCCCAUCGUUGUUGGCUCGGACUAUCCAU .......((...(((((((((.............)))))..............((((((((.(((((((((........)))))))))............)))))))).))))...)).. ( -33.94) >DroPse_CAF1 4167 120 - 1 CCGUAGAGUUGUGUCCGCUCUCUAACCACUAUAUGCAGUAUAUCCAAGAUUUCCGCGAGCAUCCGGCAGCGUAUUUGCUAGCUGCAGGCUACCCCAUCGUUAUGGGCUCCGAUUAUCCCC ...(((((..((....)).))))).......................(((...((.((((..((.(((((..........))))).)).....((((....))))))))))...)))... ( -27.30) >DroSim_CAF1 1707 120 - 1 CCGUGGAGAUCUGUCCCCUGUCCAACCAUUAUCUGCAAUACUUUAACGAUUUUCGUCAACAUCCGGCGGCGUAUCUGAUUGCCGCCGGAUUUCCAAUUGUAAUUGGGUCGGACUAUCCGU ....(((..((((.(((.((......)).....((((((......(((.....)))....(((((((((((........))))))))))).....))))))...))).))))...))).. ( -38.80) >DroEre_CAF1 16084 120 - 1 CCGUGGAGAUCUGUCCGCUGUCCAACCAUUAUCUGCAAUACUUUAACGACUUUCGUCAACAUCCGGCGGCGUAUCUGAUUGCCGCCGGUUUUCCAAUUGUAAUUGGGUCGGAUUAUCCGU ..(((((......))))).((((.(((...((.((((((........(((....))).....(((((((((........))))))))).......))))))))..))).))))....... ( -39.20) >DroYak_CAF1 16390 120 - 1 CCGUGGAGAUCUGUCCCCUGUCCAACCAUUAUCUGCAAUACUUCAACGAUUUCCGCCAACAUCCGGCGGCGUAUCUGAUUGCCGCCGGCUUUCCAAUUGUAAUUGGGUCGGACUAUCCGU ..(((((((((....................................)))))))))......(((((((((........)))))))))...((((((....)))))).(((.....))). ( -35.32) >consensus CCGUGGAGAUCUGUCCGCUGUCCAACCAUUAUCUGCAAUACUUUAACGAUUUUCGUCAACAUCCGGCGGCGUAUCUGAUUGCCGCCGGCUUUCCAAUUGUAAUUGGGUCGGACUAUCCGU ....(((.....((((..............................................(((((((((........)))))))))....(((((....)))))...))))..))).. (-26.33 = -26.28 + -0.05)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:39 2006