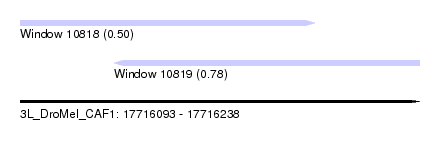
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,716,093 – 17,716,238 |
| Length | 145 |
| Max. P | 0.781560 |

| Location | 17,716,093 – 17,716,200 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 83.64 |
| Mean single sequence MFE | -30.18 |
| Consensus MFE | -19.96 |
| Energy contribution | -23.27 |
| Covariance contribution | 3.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17716093 107 + 23771897 CAACACCGCAUUAAAUGUGCACGACAUCAAAUUGAUGAGGCUGCCACGGCCGGCGGCAGAUAGAGCACCAUGUAUGCCGAUUUGGCUUUAUUUAGACCAAACUGGCU .......((((.....((((....((((.....))))..((((((......)))))).......)))).))))..((((.(((((((......)).))))).)))). ( -33.20) >DroSec_CAF1 93846 107 + 1 CAACACCACAUUAAAUGUGCACGACAUCAAAUUGAUGAGGCUGCCACGGCCGGCGGCAGAUAGAGCACCAUGGAUGCCGAUUUGGCUUUAUUUAGACCAAACUGGCU .........(((....((((....((((.....))))..((((((......)))))).......))))....)))((((.(((((((......)).))))).)))). ( -32.70) >DroSim_CAF1 87942 107 + 1 CAACACCACAUUAAAUGUGCACGACAUCAAAUUGAUGAGGCUGCCACGGCCGGCGGCAGAUAGAGCACCAUGGAUGCCGAUUUGGCUUUAUUUAGACCAAACUGGCU .........(((....((((....((((.....))))..((((((......)))))).......))))....)))((((.(((((((......)).))))).)))). ( -32.70) >DroAna_CAF1 83065 92 + 1 CAACACCACAUUAAAUGUGCACGACAUCAAAUUGAUGAGGCUGCCACUGGUUCUGGCAGAUAUGGCAAUAGUGCUGGCUAUUUAGCUAAAUU--------------- ..................((((..((((.....))))...((((((.......))))))...........))))(((((....)))))....--------------- ( -22.10) >consensus CAACACCACAUUAAAUGUGCACGACAUCAAAUUGAUGAGGCUGCCACGGCCGGCGGCAGAUAGAGCACCAUGGAUGCCGAUUUGGCUUUAUUUAGACCAAACUGGCU ................((((....((((.....))))..((((((......)))))).......)))).......((((.(((((((......)).))))).)))). (-19.96 = -23.27 + 3.31)

| Location | 17,716,127 – 17,716,238 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 68.96 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -17.67 |
| Energy contribution | -20.55 |
| Covariance contribution | 2.88 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17716127 111 - 23771897 CGACUUCUUAGAA--AAUAUGUAAGAGGACCUGGCUAACUAGCCAGUUUGGUCUAAAUAAAGCCAAAUCGGCAUACAUGGUGCUCUAUCUGCCGCCGGCCGUGGCAGCCUCAU ((((((((((.(.--....).)))))))..((((((....))))))(((((.((......))))))))))((((.....)))).....(((((((.....)))))))...... ( -34.30) >DroSec_CAF1 93880 104 - 1 --------CAGAAAGAGGUUGUAAGAGGUCC-GACUAACUAGCCAGUUUGGUCUAAAUAAAGCCAAAUCGGCAUCCAUGGUGCUCUAUCUGCCGCCGGCCGUGGCAGCCUCAU --------......((((((((....((((.-.........(((.((((((.((......)))))))).)))......(((((.......).))))))))...)))))))).. ( -37.50) >DroSim_CAF1 87976 111 - 1 CAACUUCUCAGAA--AAUUUGUAAGAGAACCUGGCUAGCUAGCCAGUUUGGUCUAAAUAAAGCCAAAUCGGCAUCCAUGGUGCUCUAUCUGCCGCCGGCCGUGGCAGCCUCAU ..........((.--........((((.((((((.......(((.((((((.((......)))))))).)))..))).))).))))..(((((((.....)))))))..)).. ( -32.00) >DroAna_CAF1 83099 90 - 1 CUAUU--CUAUAA--GAUCUUUGAGAAGAACUCUAU-------------------AAUUUAGCUAAAUAGCCAGCACUAUUGCCAUAUCUGCCAGAACCAGUGGCAGCCUCAU .....--......--......((((...........-------------------......((..(((((......))))))).....((((((.......)))))).)))). ( -13.70) >consensus C_ACU__UCAGAA__AAUUUGUAAGAGGACCUGGCUAACUAGCCAGUUUGGUCUAAAUAAAGCCAAAUCGGCAUCCAUGGUGCUCUAUCUGCCGCCGGCCGUGGCAGCCUCAU ........................((((.....((((....(((.(((((((.........))))))).))).....)))).......(((((((.....))))))))))).. (-17.67 = -20.55 + 2.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:35 2006